Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T10383
(Former ID: TTDI03468)
|
|||||
| Target Name |
Phospholipase D2 (PLD2)
|
|||||
| Synonyms |
hPLD2; Phosphatidylcholine-hydrolyzing phospholipase D2; PLD1C; PLD 2; Choline phosphatase 2
Click to Show/Hide
|
|||||
| Gene Name |
PLD2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Coagulation defect [ICD-11: 3B10] | |||||
| Function |
May have a role in signal-induced cytoskeletal regulation and/or endocytosis.
Click to Show/Hide
|
|||||
| BioChemical Class |
Phosphoric diester hydrolase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.1.4.4
|
|||||
| Sequence |
MTATPESLFPTGDELDSSQLQMESDEVDTLKEGEDPADRMHPFLAIYELQSLKVHPLVFA
PGVPVTAQVVGTERYTSGSKVGTCTLYSVRLTHGDFSWTTKKKYRHFQELHRDLLRHKVL MSLLPLARFAVAYSPARDAGNREMPSLPRAGPEGSTRHAASKQKYLENYLNRLLTMSFYR NYHAMTEFLEVSQLSFIPDLGRKGLEGMIRKRSGGHRVPGLTCCGRDQVCYRWSKRWLVV KDSFLLYMCLETGAISFVQLFDPGFEVQVGKRSTEARHGVRIDTSHRSLILKCSSYRQAR WWAQEITELAQGPGRDFLQLHRHDSYAPPRPGTLARWFVNGAGYFAAVADAILRAQEEIF ITDWWLSPEVYLKRPAHSDDWRLDIMLKRKAEEGVRVSILLFKEVELALGINSGYSKRAL MLLHPNIKVMRHPDQVTLWAHHEKLLVVDQVVAFLGGLDLAYGRWDDLHYRLTDLGDSSE SAASQPPTPRPDSPATPDLSHNQFFWLGKDYSNLITKDWVQLDRPFEDFIDRETTPRMPW RDVGVVVHGLPARDLARHFIQRWNFTKTTKAKYKTPTYPYLLPKSTSTANQLPFTLPGGQ CTTVQVLRSVDRWSAGTLENSILNAYLHTIRESQHFLYIENQFFISCSDGRTVLNKVGDE IVDRILKAHKQGWCYRVYVLLPLLPGFEGDISTGGGNSIQAILHFTYRTLCRGEYSILHR LKAAMGTAWRDYISICGLRTHGELGGHPVSELIYIHSKVLIADDRTVIIGSANINDRSLL GKRDSELAVLIEDTETEPSLMNGAEYQAGRFALSLRKHCFGVILGANTRPDLDLRDPICD DFFQLWQDMAESNANIYEQIFRCLPSNATRSLRTLREYVAVEPLATVSPPLARSELTQVQ GHLVHFPLKFLEDESLLPPLGSKEGMIPLEVWT Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | VU0364739 | Drug Info | Clinical trial | Coagulation defect | [2] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | VU0364739 | Drug Info | [1] | |||
| Activator | [+] 1 Activator drugs | + | ||||
| 1 | PIP2 | Drug Info | [3] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Halopemide | Ligand Info | |||||
| Structure Description | Structure of compound 1 (halopemide) bound human Phospholipase D2 catalytic domain | PDB:6OHP | ||||
| Method | X-ray diffraction | Resolution | 2.60 Å | Mutation | No | [4] |
| PDB Sequence |
FLQLHRHDSY
326 APPRPGTLAR336 WFVNGAGYFA346 AVADAILRAQ356 EEIFITDWWL366 SPEVYLKRPA 376 HSDDWRLDIM386 LKRKAEEGVR396 VSILLFKEVE406 LALGINSGYS416 KRALMLLHPN 426 IKVMRHPDQV436 TLWAHHEKLL446 VVDQVVAFLG456 GLDLAYGRWD466 DLHYRLTDLG 476 DLSHNQFFWL507 GKDYSNLITK517 DWVQLDRPFE527 DFIDRETTPR537 MPWRDVGVVV 547 HGLPARDLAR557 HFIQRWNFTK567 TTKAKKTPTY578 PYLLPKSTST588 FTLPGGQCTT 603 VQVLRSVDRW613 SAGTLENSIL623 NAYLHTIRES633 QHFLYIENQF643 FISCSDGRTV 653 LNKVGDEIVD663 RILKAHKQGW673 CYRVYVLLPL683 LPGFEGDIST693 GGGNSIQAIL 703 HFTYRTLCRG713 EYSILHRLKA723 AMGTAWRDYI733 SICGLRTHGE743 LGGHPVSELI 753 YIHSKVLIAD763 DRTVIIGSAN773 INDRSLLGKR783 DSELAVLIED793 TETEPSLMNG 803 AEYQAGRFAL813 SLRKHCFGVI823 LGANTRPDLD833 LRDPICDDFF843 QLWQDMAESN 853 ANIYEQIFRC863 LPSNATRSLR873 TLREYVAVEP883 LATVSPPLAR893 SELTQVQGHL 903 VHFPLKFLED913 ESLLPPGMIP928 LEVWT
|
|||||
|
|
TRP364
3.458
TRP365
3.326
PHE402
4.652
LEU409
3.623
GLY410
3.187
ILE411
3.653
LEU438
3.921
TRP439
4.937
ALA440
3.624
HIS442
3.218
ARG464
2.484
LEU514
3.430
ASP518
3.667
|
|||||
| Ligand Name: Phosphonotyrosine | Ligand Info | |||||
| Structure Description | Structure of compound 1 (halopemide) bound human Phospholipase D2 catalytic domain | PDB:6OHP | ||||
| Method | X-ray diffraction | Resolution | 2.60 Å | Mutation | No | [4] |
| PDB Sequence |
FLQLHRHDSY
326 APPRPGTLAR336 WFVNGAGYFA346 AVADAILRAQ356 EEIFITDWWL366 SPEVYLKRPA 376 HSDDWRLDIM386 LKRKAEEGVR396 VSILLFKEVE406 LALGINSGYS416 KRALMLLHPN 426 IKVMRHPDQV436 TLWAHHEKLL446 VVDQVVAFLG456 GLDLAYGRWD466 DLHYRLTDLG 476 DLSHNQFFWL507 GKDYSNLITK517 DWVQLDRPFE527 DFIDRETTPR537 MPWRDVGVVV 547 HGLPARDLAR557 HFIQRWNFTK567 TTKAKKTPTY578 PYLLPKSTST588 FTLPGGQCTT 603 VQVLRSVDRW613 SAGTLENSIL623 NAYLHTIRES633 QHFLYIENQF643 FISCSDGRTV 653 LNKVGDEIVD663 RILKAHKQGW673 CYRVYVLLPL683 LPGFEGDIST693 GGGNSIQAIL 703 HFTYRTLCRG713 EYSILHRLKA723 AMGTAWRDYI733 SICGLRTHGE743 LGGHPVSELI 753 YIHSKVLIAD763 DRTVIIGSAN773 INDRSLLGKR783 DSELAVLIED793 TETEPSLMNG 803 AEYQAGRFAL813 SLRKHCFGVI823 LGANTRPDLD833 LRDPICDDFF843 QLWQDMAESN 853 ANIYEQIFRC863 LPSNATRSLR873 TLREYVAVEP883 LATVSPPLAR893 SELTQVQGHL 903 VHFPLKFLED913 ESLLPPGMIP928 LEVWT
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|



| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Glycerophospholipid metabolism | hsa00564 | Affiliated Target |

|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
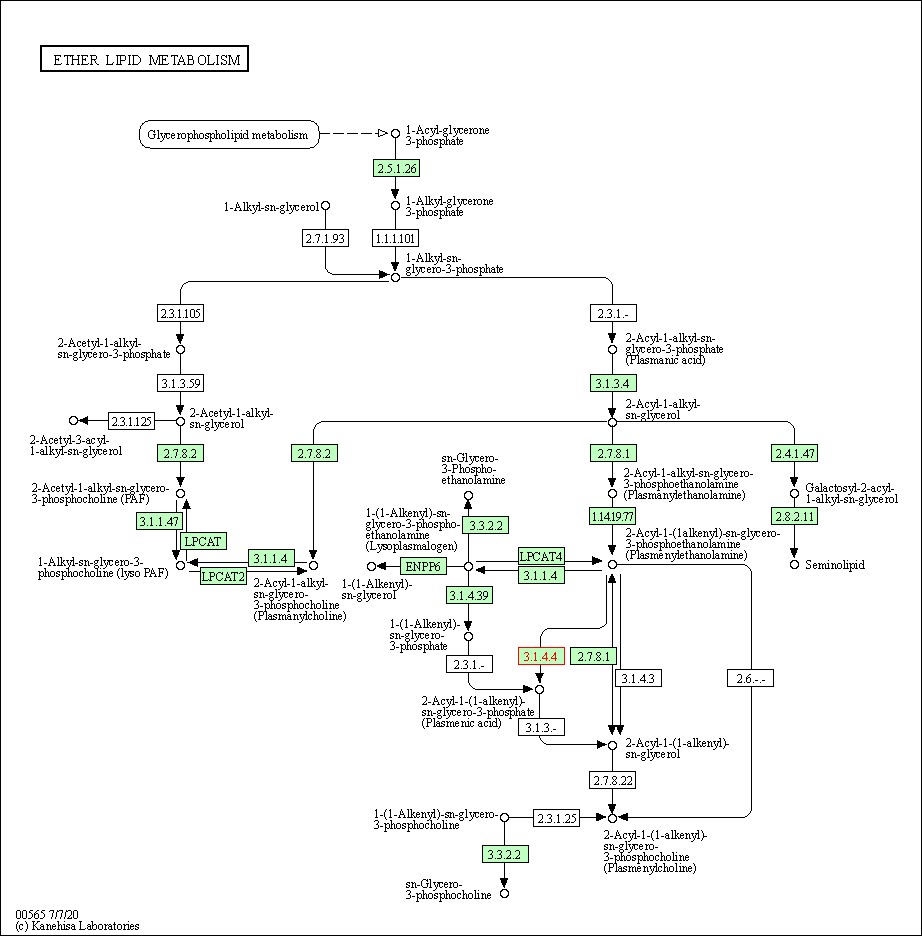
| Ether lipid metabolism | hsa00565 | Affiliated Target |

|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
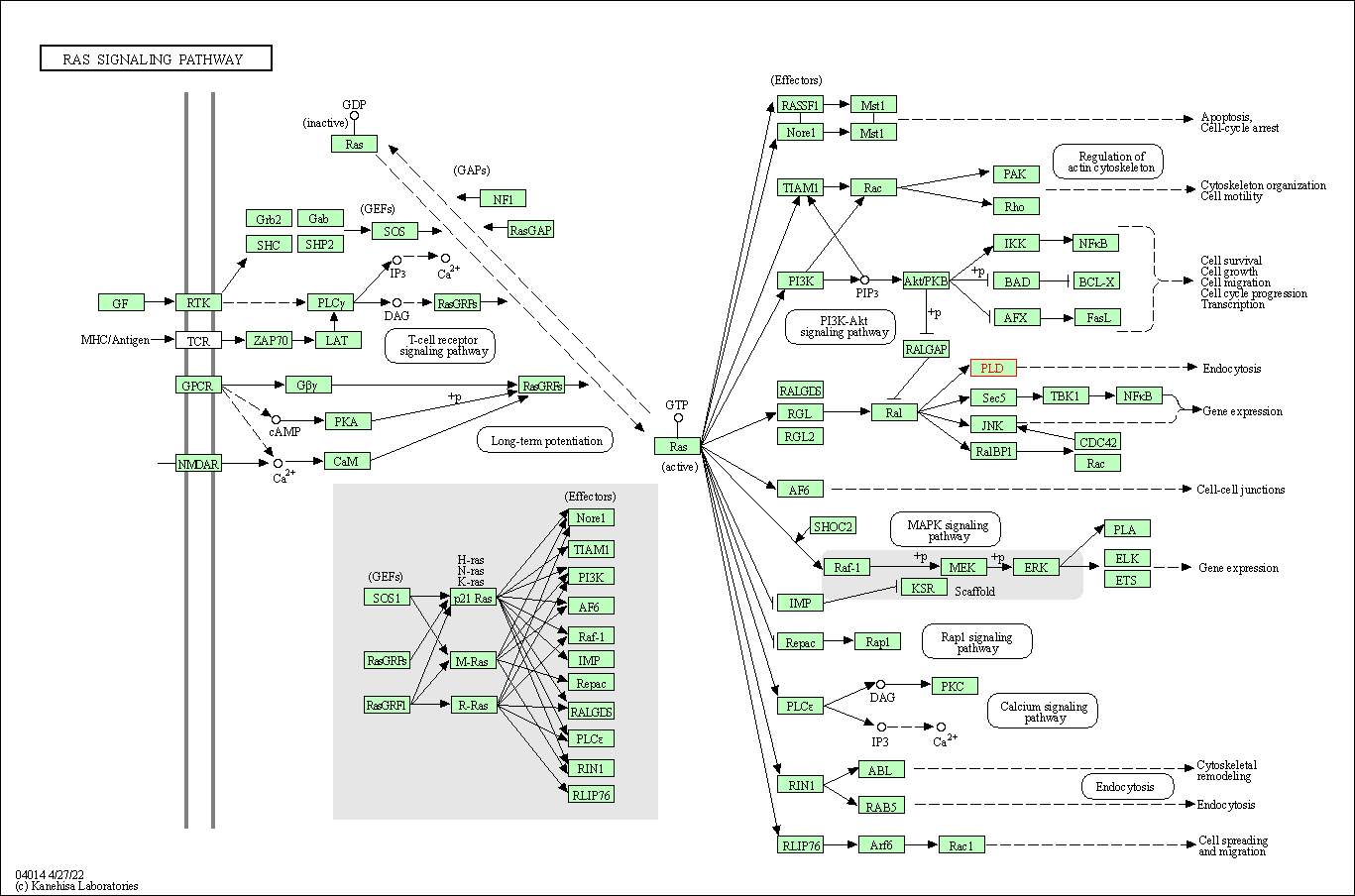
| Ras signaling pathway | hsa04014 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
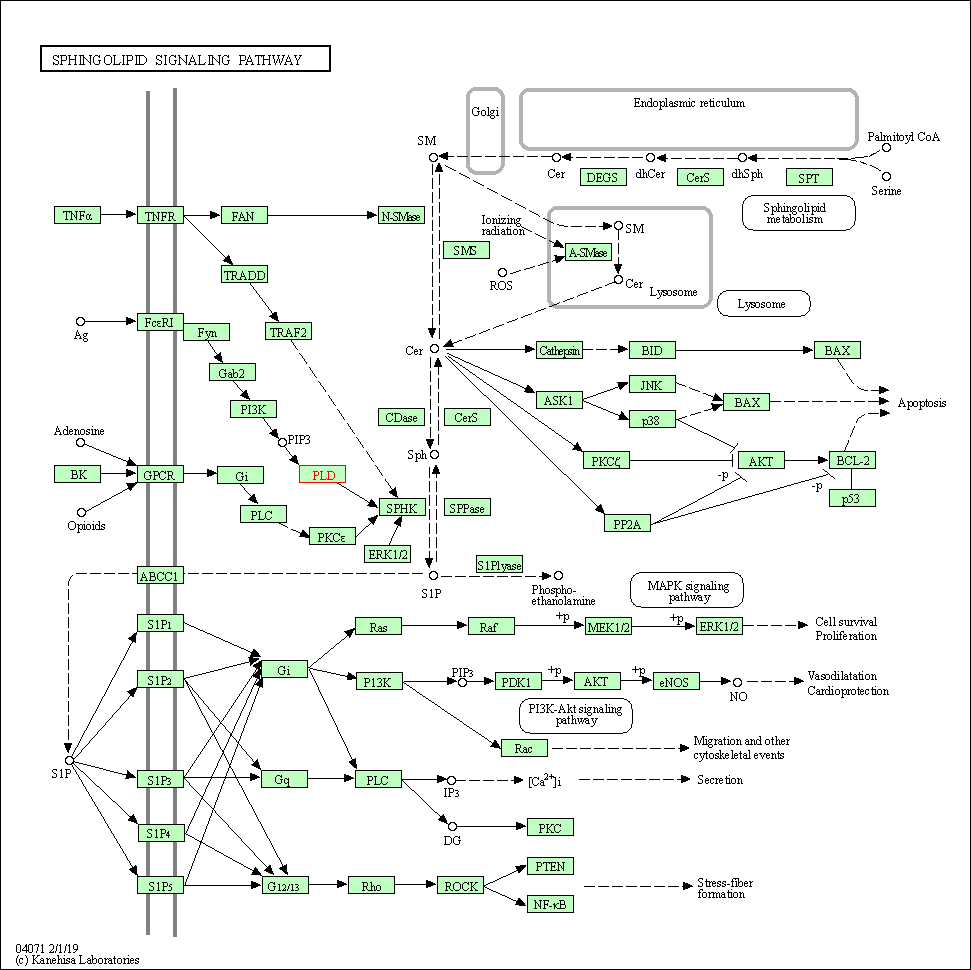
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
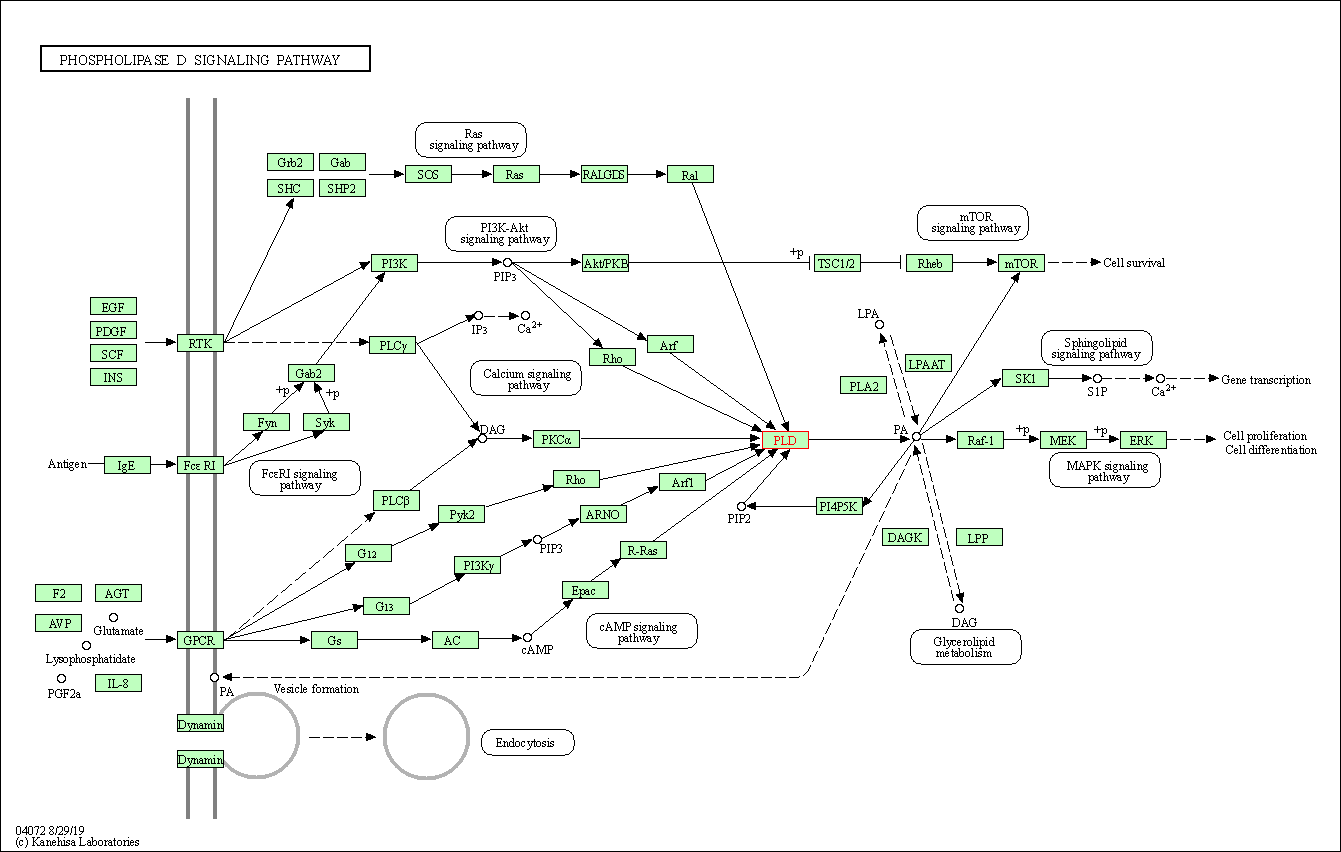
| Phospholipase D signaling pathway | hsa04072 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Endocytosis | hsa04144 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
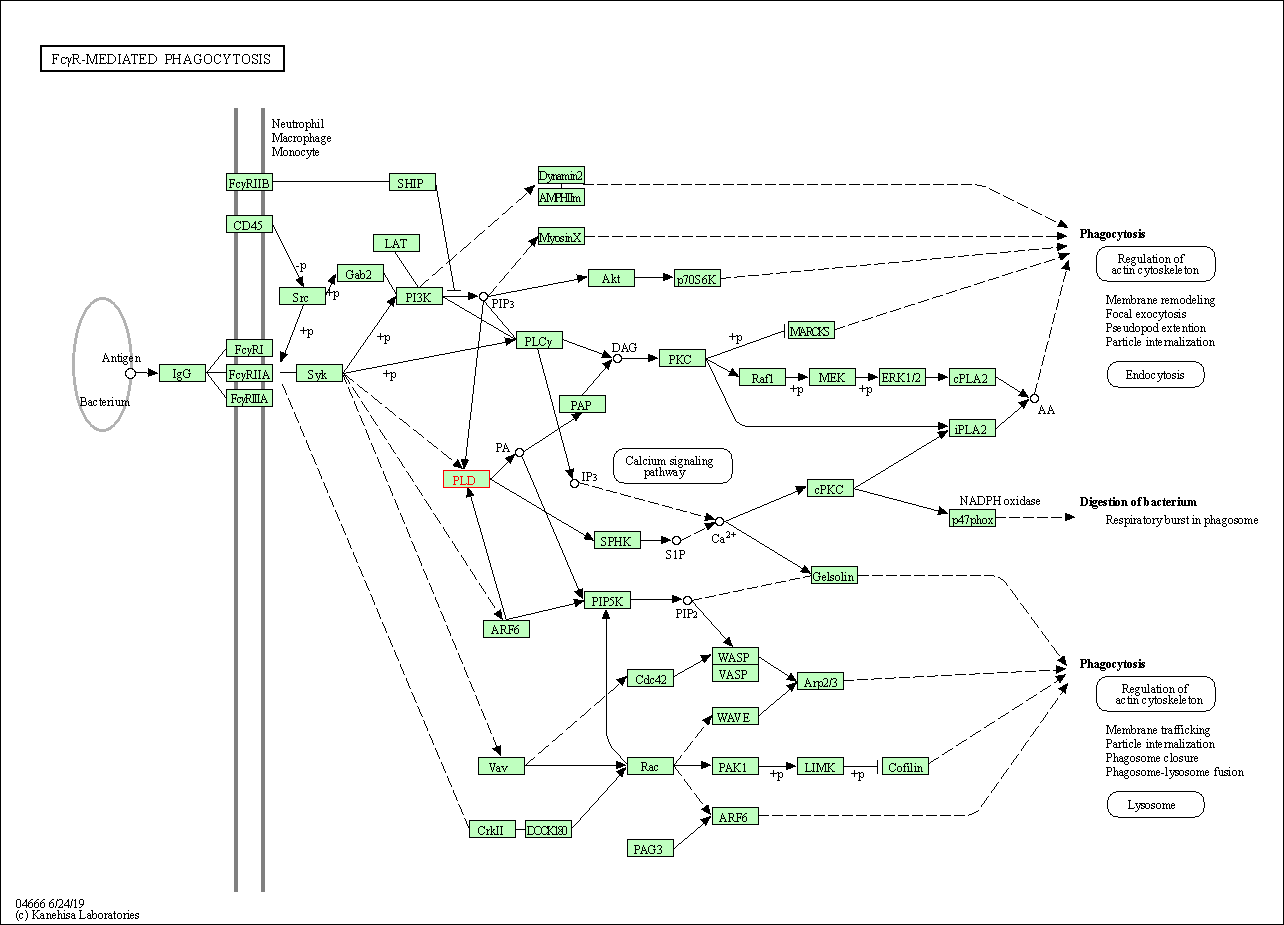
| Fc gamma R-mediated phagocytosis | hsa04666 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
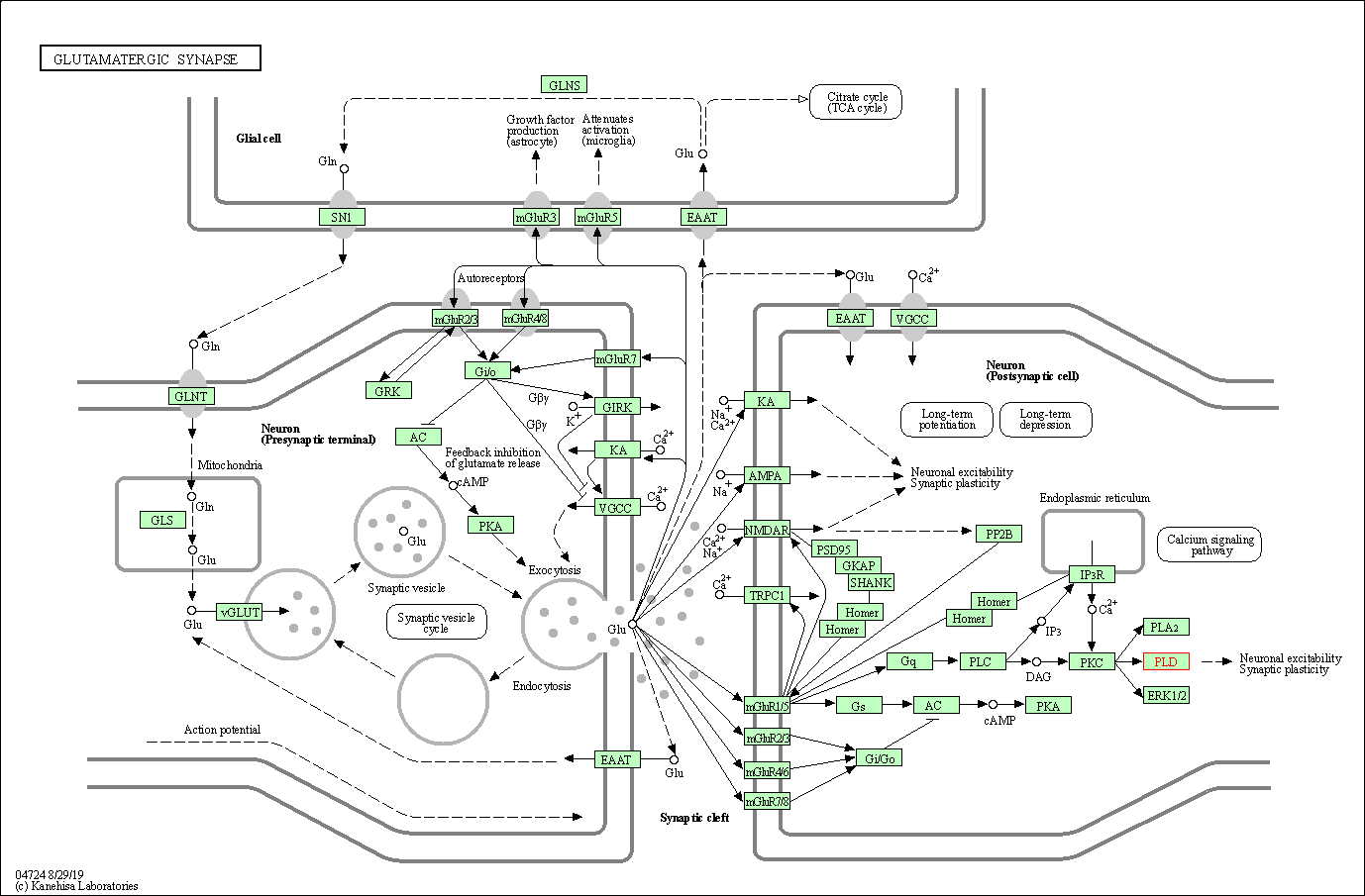
| Glutamatergic synapse | hsa04724 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
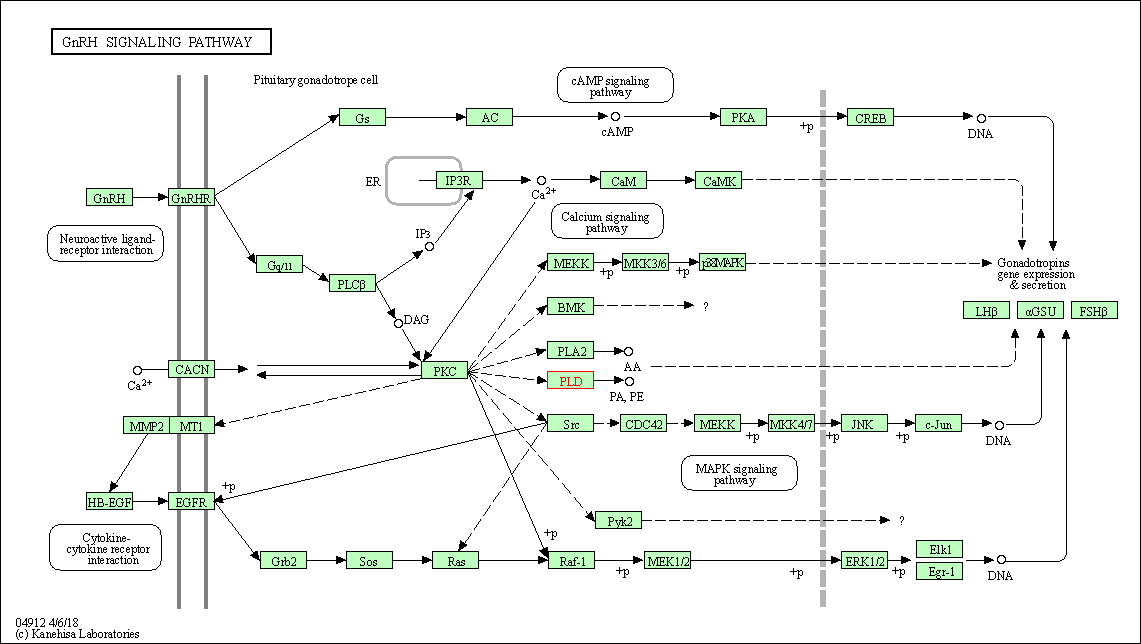
| GnRH signaling pathway | hsa04912 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
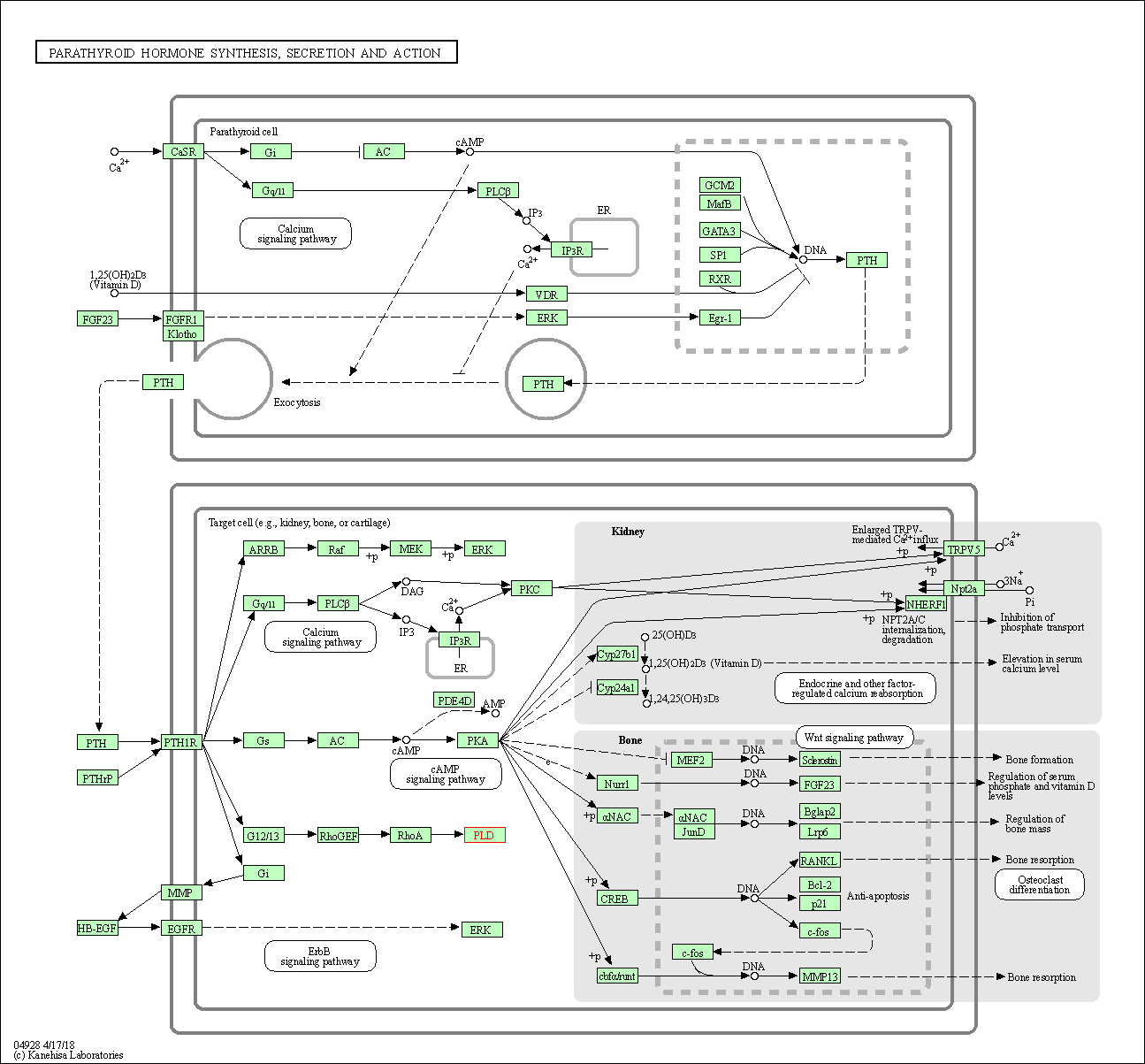
| Parathyroid hormone synthesis, secretion and action | hsa04928 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 8 | Degree centrality | 8.59E-04 | Betweenness centrality | 8.94E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.32E-01 | Radiality | 1.41E+01 | Clustering coefficient | 1.07E-01 |
| Neighborhood connectivity | 5.33E+01 | Topological coefficient | 1.46E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Design, synthesis, and biological evaluation of halogenated N-(2-(4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-8-yl)ethyl)benzamides: discovery of an isoform-selective small molecule phospholipase D2 inhibitor. J Med Chem. 2010 Sep 23;53(18):6706-19. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5287). | |||||
| REF 3 | Cloning and initial characterization of a human phospholipase D2 (hPLD2). ADP-ribosylation factor regulates hPLD2. J Biol Chem. 1998 May 22;273(21):12846-52. | |||||
| REF 4 | Human PLD structures enable drug design and characterization of isoenzyme selectivity. Nat Chem Biol. 2020 Apr;16(4):391-399. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

