Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T53904
(Former ID: TTDC00054)
|
|||||
| Target Name |
HIF-prolyl hydroxylase 1 (HPH-1)
|
|||||
| Synonyms |
Prolyl hydroxylase domain-containing protein 1; PHD1; Hypoxia-inducible factor prolyl hydroxylase 1; HPH-3; HIF-PH1; HIF-PH; Estrogen-induced tag6; Estrogen-induced tag 6; Egl nine homolog 2; EIT6; EIT-6
Click to Show/Hide
|
|||||
| Gene Name |
EGLN2
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Acute disease anaemia [ICD-11: 3A90] | |||||
| Function |
Hydroxylates a specific proline found in each of the oxygen-dependent degradation (ODD) domains (N-terminal, NODD, and C-terminal, CODD) of HIF1A. Also hydroxylates HIF2A. Has a preference for the CODD site for both HIF1A and HIF2A. Hydroxylated HIFs are then targeted for proteasomal degradation via the von Hippel-Lindau ubiquitination complex. Under hypoxic conditions, the hydroxylation reaction is attenuated allowing HIFs to escape degradation resulting in their translocation to the nucleus, heterodimerization with HIF1B, and increased expression of hypoxy-inducible genes. EGLN2 is involved in regulating hypoxia tolerance and apoptosis in cardiac and skeletal muscle. Also regulates susceptibility to normoxic oxidative neuronal death. Links oxygen sensing to cell cycle and primary cilia formation by hydroxylating the critical centrosome component CEP192 which promotes its ubiquitination and subsequent proteasomal degradation. Hydroxylates IKBKB, mediating NF-kappaB activation in hypoxic conditions. Target proteins are preferentially recognized via a LXXLAP motif. Cellular oxygen sensor that catalyzes, under normoxic conditions, the post-translational formation of 4-hydroxyproline in hypoxia-inducible factor (HIF) alpha proteins.
Click to Show/Hide
|
|||||
| BioChemical Class |
Paired donor oxygen oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.14.11.29
|
|||||
| Sequence |
MDSPCQPQPLSQALPQLPGSSSEPLEPEPGRARMGVESYLPCPLLPSYHCPGVPSEASAG
SGTPRATATSTTASPLRDGFGGQDGGELRPLQSEGAAALVTKGCQRLAAQGARPEAPKRK WAEDGGDAPSPSKRPWARQENQEAEREGGMSCSCSSGSGEASAGLMEEALPSAPERLALD YIVPCMRYYGICVKDSFLGAALGGRVLAEVEALKRGGRLRDGQLVSQRAIPPRSIRGDQI AWVEGHEPGCRSIGALMAHVDAVIRHCAGRLGSYVINGRTKAMVACYPGNGLGYVRHVDN PHGDGRCITCIYYLNQNWDVKVHGGLLQIFPEGRPVVANIEPLFDRLLIFWSDRRNPHEV KPAYATRYAITVWYFDAKERAAAKDKYQLASGQKGVQVPVSQPPTPT Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Daprodustat | Drug Info | Approved | Anaemia | [2] | |
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | FG-4592 | Drug Info | Phase 3 | Kidney disease | [3], [4] | |
| 2 | BAY-85-3934 | Drug Info | Phase 2 | Anemia | [5], [6] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 5 Inhibitor drugs | + | ||||
| 1 | Daprodustat | Drug Info | [7] | |||
| 2 | FG-4592 | Drug Info | [1] | |||
| 3 | BAY-85-3934 | Drug Info | [8] | |||
| 4 | 2-(carboxymethylamino)-2-oxoacetic acid | Drug Info | [9] | |||
| 5 | Pyridine-2,4-dicarboxylic acid | Drug Info | [9] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Phd-1-IN-1 | Ligand Info | |||||
| Structure Description | Structure of PHD1 in complex with 1,2,4-Triazolo-[1,5-a]pyridine | PDB:5V1B | ||||
| Method | X-ray diffraction | Resolution | 2.49 Å | Mutation | No | [10] |
| PDB Sequence |
PSAPERLALD
180 YIVPCMRYYG190 ICVVDSFLGA200 ALGGRVLAEV210 EALKRGGRLR220 QLVSPRSIRG 237 DQIAWVEGHE247 PGCRSIGALM257 AHVDAVIRHC267 AGRLGSYKIN277 GRTKAMVACY 287 PGNGLGYVRH297 VDNPHGDGRC307 ITCIYYLNQN317 WDVKVHGGLL327 QIFPEGRPVV 337 ANIEPLFDRL347 LIFWSDRRNP357 HEVKPAYATR367 YAITVWYFDA377 DERARAKDKY 387 QLASGQKGVQ397 VPVSQP
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
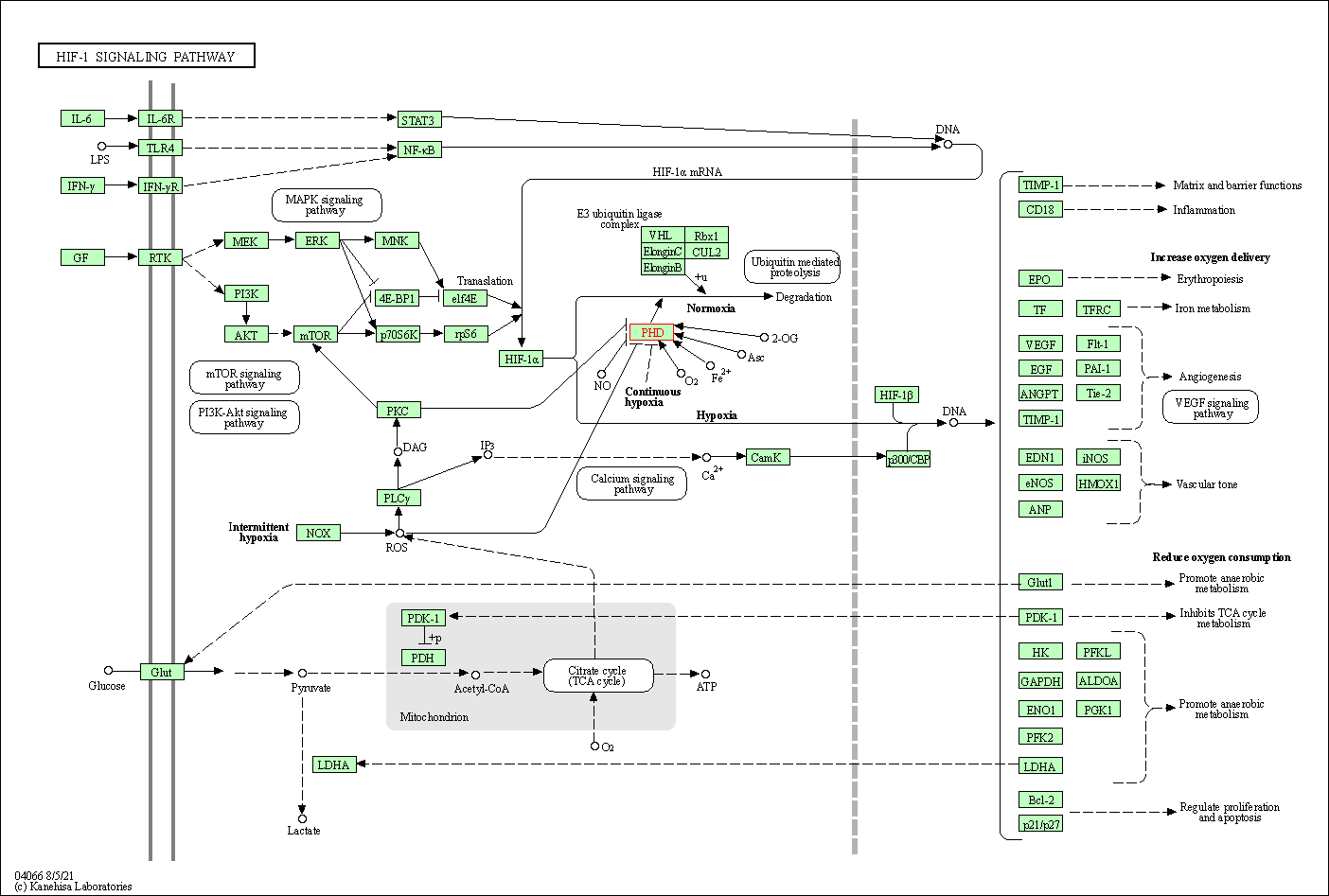
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| HIF-1 signaling pathway | hsa04066 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 2.13E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.20E-01 | Radiality | 1.39E+01 | Clustering coefficient | 1.67E-01 |
| Neighborhood connectivity | 3.13E+01 | Topological coefficient | 2.87E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | HIF-1 signaling pathway | |||||
| 2 | Pathways in cancer | |||||
| 3 | Renal cell carcinoma | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Hypoxia response via HIF activation | |||||
| PID Pathway | [+] 2 PID Pathways | + | ||||
| 1 | HIF-2-alpha transcription factor network | |||||
| 2 | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | The latest advances in kidney diseases and related disorders. Drug News Perspect. 2007 Dec;20(10):647-54. | |||||
| REF 2 | FDA Approved Drug Products from FDA Official Website. 2023. Application Number: 216951. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 8454). | |||||
| REF 4 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800023523) | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 8456). | |||||
| REF 6 | ClinicalTrials.gov (NCT01975818) Maintenance Treatment of Anemia Associated With Chronic Kidney Disease (CKD) in Hemodialysis Subjects on Epoetin Alfa / Beta Treatment Versus BAY85-3934. U.S. National Institutes of Health. | |||||
| REF 7 | Clinical pipeline report, company report or official report of GlaxoSmithKline (2009). | |||||
| REF 8 | Mimicking hypoxia to treat anemia: HIF-stabilizer BAY 85-3934 (Molidustat) stimulates erythropoietin production without hypertensive effects. PLoS One. 2014 Nov 13;9(11):e111838. | |||||
| REF 9 | Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone de... J Med Chem. 2010 Aug 12;53(15):5629-38. | |||||
| REF 10 | 1,2,4-Triazolo-[1,5-a]pyridine HIF Prolylhydroxylase Domain-1 (PHD-1) Inhibitors With a Novel Monodentate Binding Interaction. J Med Chem. 2017 Jul 13;60(13):5663-5672. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

