Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T06093
(Former ID: TTDI02162)
|
|||||
| Target Name |
Rho-associated protein kinase 2 (ROCK2)
|
|||||
| Synonyms |
p164 ROCK-2; Rho-associated, coiled-coil-containing protein kinase II; Rho-associated, coiled-coil-containing protein kinase 2; Rho kinase 2; ROCK-II; KIAA0619
Click to Show/Hide
|
|||||
| Gene Name |
ROCK2
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Graft-versus-host disease [ICD-11: 4B24] | |||||
| Function |
Involved in regulation of smooth muscle contraction, actin cytoskeleton organization, stress fiber and focal adhesion formation, neurite retraction, cell adhesion and motility via phosphorylation of ADD1, BRCA2, CNN1, EZR, DPYSL2, EP300, MSN, MYL9/MLC2, NPM1, RDX, PPP1R12A and VIM. Phosphorylates SORL1 and IRF4. Acts as a negative regulator of VEGF-induced angiogenic endothelial cell activation. Positively regulates the activation of p42/MAPK1-p44/MAPK3 and of p90RSK/RPS6KA1 during myogenic differentiation. Plays an important role in the timely initiation of centrosome duplication. Inhibits keratinocyte terminal differentiation. May regulate closure of the eyelids and ventral body wall through organization of actomyosin bundles. Plays a critical role in the regulation of spine and synaptic properties in the hippocampus. Plays an important role in generating the circadian rhythm of the aortic myofilament Ca(2+) sensitivity and vascular contractility by modulating the myosin light chain phosphorylation. Protein kinase which is a key regulator of actin cytoskeleton and cell polarity.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MSRPPPTGKMPGAPETAPGDGAGASRQRKLEALIRDPRSPINVESLLDGLNSLVLDLDFP
ALRKNKNIDNFLNRYEKIVKKIRGLQMKAEDYDVVKVIGRGAFGEVQLVRHKASQKVYAM KLLSKFEMIKRSDSAFFWEERDIMAFANSPWVVQLFYAFQDDRYLYMVMEYMPGGDLVNL MSNYDVPEKWAKFYTAEVVLALDAIHSMGLIHRDVKPDNMLLDKHGHLKLADFGTCMKMD ETGMVHCDTAVGTPDYISPEVLKSQGGDGFYGRECDWWSVGVFLYEMLVGDTPFYADSLV GTYSKIMDHKNSLCFPEDAEISKHAKNLICAFLTDREVRLGRNGVEEIRQHPFFKNDQWH WDNIRETAAPVVPELSSDIDSSNFDDIEDDKGDVETFPIPKAFVGNQLPFIGFTYYRENL LLSDSPSCRETDSIQSRKNEESQEIQKKLYTLEEHLSNEMQAKEELEQKCKSVNTRLEKT AKELEEEITLRKSVESALRQLEREKALLQHKNAEYQRKADHEADKKRNLENDVNSLKDQL EDLKKRNQNSQISTEKVNQLQRQLDETNALLRTESDTAARLRKTQAESSKQIQQLESNNR DLQDKNCLLETAKLKLEKEFINLQSALESERRDRTHGSEIINDLQGRICGLEEDLKNGKI LLAKVELEKRQLQERFTDLEKEKSNMEIDMTYQLKVIQQSLEQEEAEHKATKARLADKNK IYESIEEAKSEAMKEMEKKLLEERTLKQKVENLLLEAEKRCSLLDCDLKQSQQKINELLK QKDVLNEDVRNLTLKIEQETQKRCLTQNDLKMQTQQVNTLKMSEKQLKQENNHLMEMKMN LEKQNAELRKERQDADGQMKELQDQLEAEQYFSTLYKTQVRELKEECEEKTKLGKELQQK KQELQDERDSLAAQLEITLTKADSEQLARSIAEEQYSDLEKEKIMKELEIKEMMARHKQE LTEKDATIASLEETNRTLTSDVANLANEKEELNNKLKDVQEQLSRLKDEEISAAAIKAQF EKQLLTERTLKTQAVNKLAEIMNRKEPVKRGNDTDVRRKEKENRKLHMELKSEREKLTQQ MIKYQKELNEMQAQIAEESQIRIELQMTLDSKDSDIEQLRSQLQALHIGLDSSSIGSGPG DAEADDGFPESRLEGWLSLPVRNNTKKFGWVKKYVIVSSKKILFYDSEQDKEQSNPYMVL DIDKLFHVRPVTQTDVYRADAKEIPRIFQILYANEGESKKEQEFPVEPVGEKSNYICHKG HEFIPTLYHFPTNCEACMKPLWHMFKPPPALECRRCHIKCHKDHMDKKEEIIAPCKVYYD ISTAKNLLLLANSTEEQQKWVSRLVKKIPKKPPAPDPFARSSPRTSMKIQQNQSIRRPSR QLAPNKPS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T12W63 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Belumosudil | Drug Info | Approved | Graft-versus-host disease | [2] | |
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | INS-117548 | Drug Info | Phase 1 | Glaucoma/ocular hypertension | [3] | |
| 2 | GSK269962A | Drug Info | Clinical trial | Inflammation | [4] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | CDE-5110 | Drug Info | Terminated | Inflammation | [5] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 25 Inhibitor drugs | + | ||||
| 1 | Belumosudil | Drug Info | [1] | |||
| 2 | GSK269962A | Drug Info | [4] | |||
| 3 | Indazolyl-thiadiazolamine derivative 1 | Drug Info | [7] | |||
| 4 | PMID28048944-Compound-11 | Drug Info | [7] | |||
| 5 | PMID28048944-Compound-19 | Drug Info | [7] | |||
| 6 | PMID28048944-Compound-2 | Drug Info | [7] | |||
| 7 | PMID28048944-Compound-3 | Drug Info | [7] | |||
| 8 | PMID28048944-Compound-4 | Drug Info | [7] | |||
| 9 | PMID28048944-Compound-5 | Drug Info | [7] | |||
| 10 | PMID28048944-Compound-6 | Drug Info | [7] | |||
| 11 | PMID28048944-Compound-7 | Drug Info | [7] | |||
| 12 | Tricyclic compound 4 | Drug Info | [7] | |||
| 13 | Tricyclic compound 5 | Drug Info | [7] | |||
| 14 | Tricyclic compound 6 | Drug Info | [7] | |||
| 15 | Tricyclic compound 7 | Drug Info | [7] | |||
| 16 | CDE-5110 | Drug Info | [8] | |||
| 17 | AMA-237 | Drug Info | [9] | |||
| 18 | ATS-907 | Drug Info | [9] | |||
| 19 | Glycyl-H 1152 | Drug Info | [10] | |||
| 20 | PMID20462760C22 | Drug Info | [11] | |||
| 21 | PMID20471253C32 | Drug Info | [12] | |||
| 22 | PMID20684608C35 | Drug Info | [13] | |||
| 23 | RKI-1447 | Drug Info | [14] | |||
| 24 | SR-3850 | Drug Info | [9] | |||
| 25 | TRN-101 | Drug Info | [9] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | INS-117548 | Drug Info | [6] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: TAK-931 | Ligand Info | |||||
| Structure Description | Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Anti-Tumor Agent | PDB:6P5P | ||||
| Method | X-ray diffraction | Resolution | 3.30 Å | Mutation | Yes | [15] |
| PDB Sequence |
QRKLEALIRD
36 PRSPINVESL46 LDGLNSLVLD56 LDFPALRKNK66 NIDNFLNRYE76 KIVKKIRGLQ 86 MKAEDYDVVK96 VIGRGAFGEV106 QLVRHKASQK116 VYAMKLLSKF126 EMIKRSDSAF 136 FWEERDIMAF146 ANSPWVVQLF156 YAFQDDRYLY166 MVMEYMPGGD176 LVNLMSNYDV 186 PEKWAKFYTA196 EVVLALDAIH206 SMGLIHRDVK216 PDNMLLDKHG226 HLKLADFGTC 236 MKMDETGMVH246 CDTAVGTPDY256 ISPEVLKSQG266 GDGYYGRECD276 WWSVGVFLYE 286 MLVGDTPFYA296 DSLVGTYSKI306 MDHKNSLCFP316 EDAEISKHAK326 NLICAFLTDR 336 EVRLGRNGVE346 EIRQHPFFKN356 DQWHWDNIRE366 TAAPVVPELS376 SDIDSSNFDD 386 IEVETFPIPK401 AFVGNQLPFI411 GFTYYR
|
|||||
|
|
||||||
| Ligand Name: N-[(2,3-dihydro-1,4-benzodioxin-5-yl)methyl]-4-(pyridin-4-yl)benzamide | Ligand Info | |||||
| Structure Description | Crystal structure of Rock2 with a pyridinylbenzamide based inhibitor | PDB:6ED6 | ||||
| Method | X-ray diffraction | Resolution | 2.86 Å | Mutation | No | [16] |
| PDB Sequence |
QRKLEALIRD
36 PRSPINVESL46 LDGLNSLVLD56 LDFPALRKNK66 NIDNFLNRYE76 KIVKKIRGLQ 86 MKAEDYDVVK96 VIGRGAFGEV106 QLVRHKASQK116 VYAMKLLSKF126 EMIKRSDSAF 136 FWEERDIMAF146 ANSPWVVQLF156 YAFQDDRYLY166 MVMEYMPGGD176 LVNLMSNYDV 186 PEKWAKFYTA196 EVVLALDAIH206 SMGLIHRDVK216 PDNMLLDKHG226 HLKLADFGTC 236 MKMDETGMVH246 CDTAVGTPDY256 ISPEVLKSQF270 YGRECDWWSV280 GVFLYEMLVG 290 DTPFYADSLV300 GTYSKIMDHK310 NSLCFPEDAE320 ISKHAKNLIC330 AFLTDREVRL 340 GRNGVEEIRQ350 HPFFKNDQWH360 WDNIRETAAP370 VVPELSSDID380 SSNFDDIETF 397 PIPKAFVGNQ407 LPFIGFTY
|
|||||
|
|
ILE98
3.572
ARG100
4.414
GLY101
3.627
ALA102
3.575
PHE103
3.054
GLY104
3.293
GLU105
3.844
VAL106
3.776
ALA119
3.598
LYS121
2.873
LEU122
3.900
LEU123
3.674
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| cGMP-PKG signaling pathway | hsa04022 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Chemokine signaling pathway | hsa04062 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
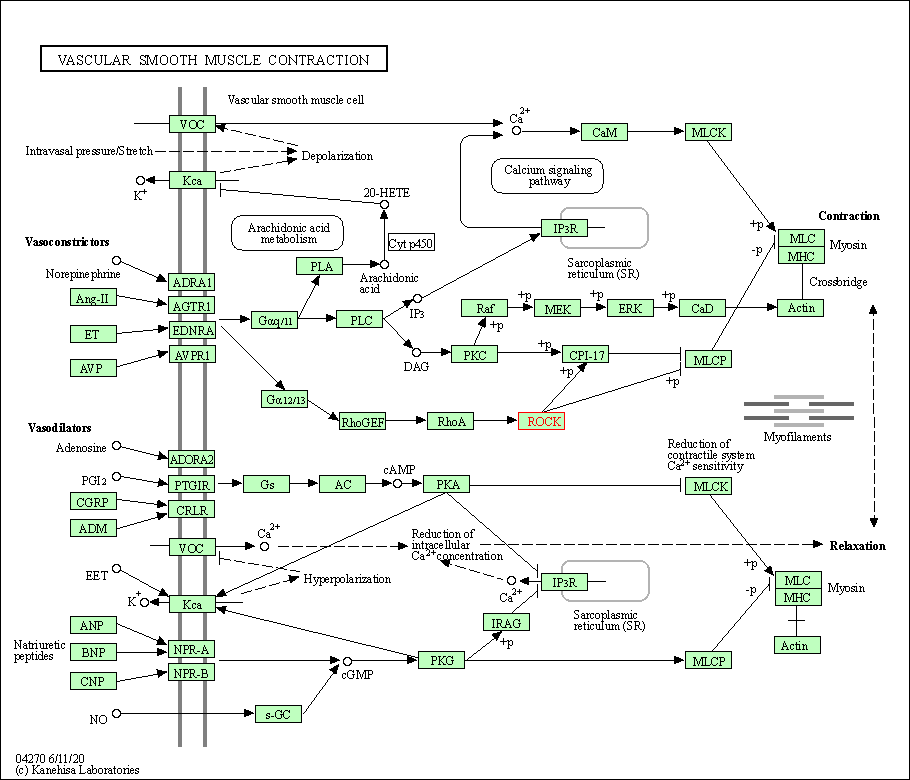
| Vascular smooth muscle contraction | hsa04270 | Affiliated Target |

|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
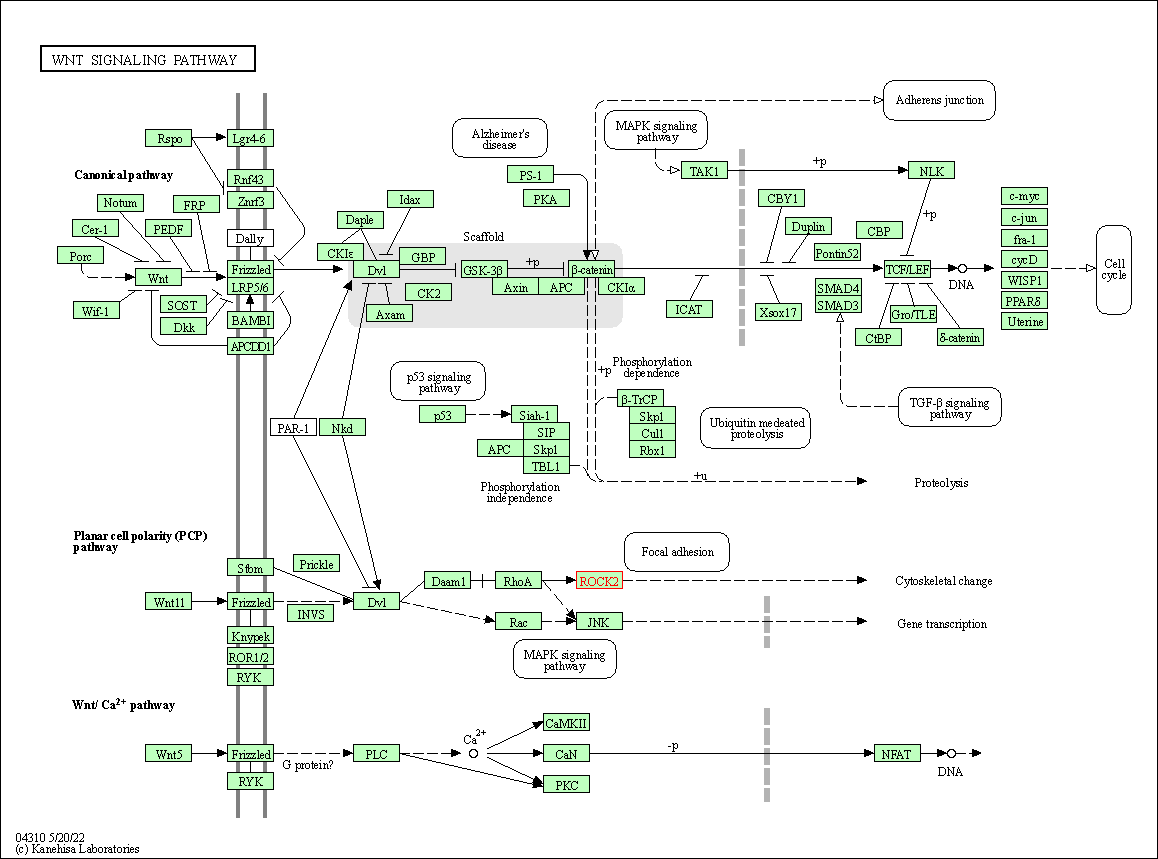
| Wnt signaling pathway | hsa04310 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
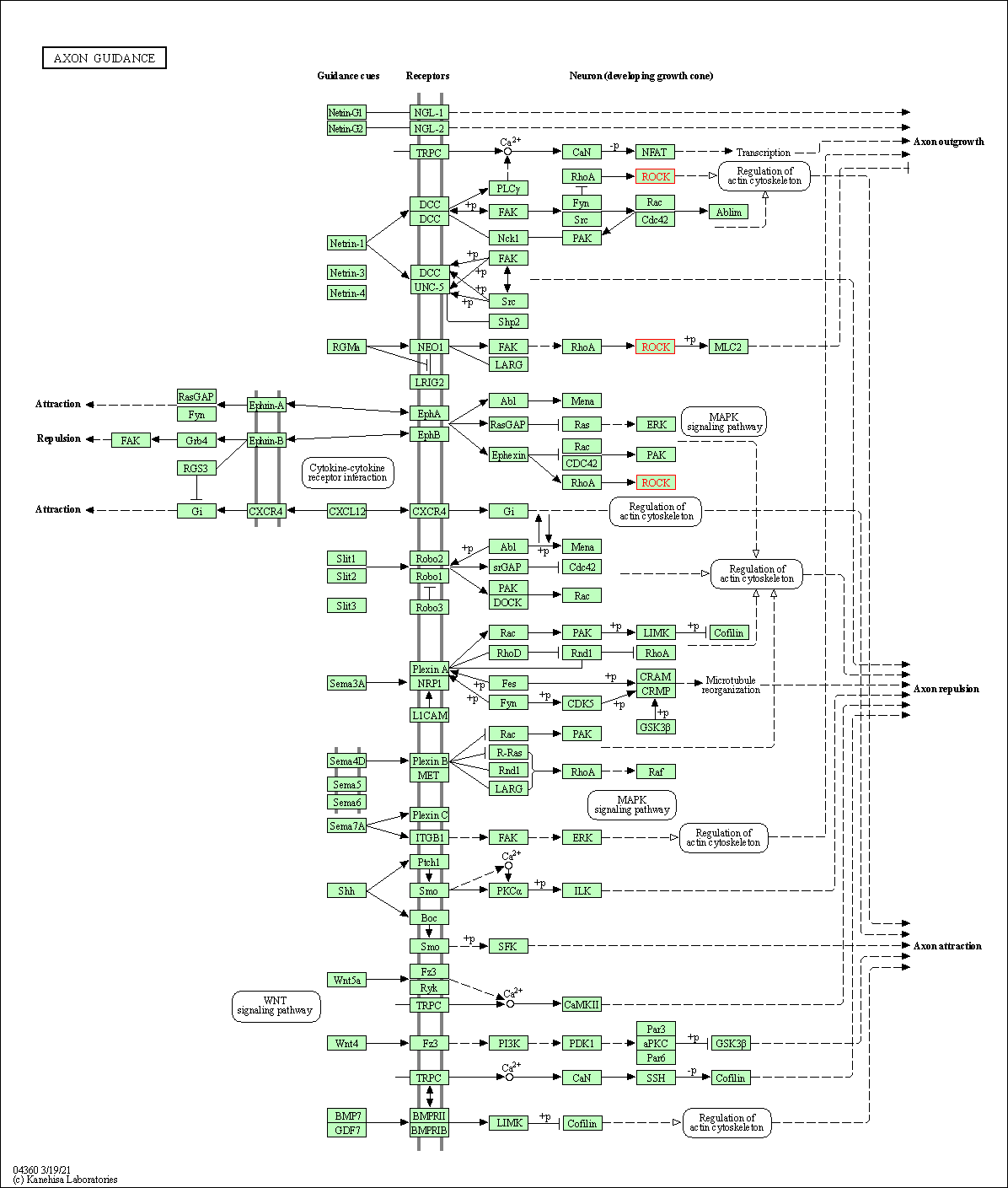
| Axon guidance | hsa04360 | Affiliated Target |

|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
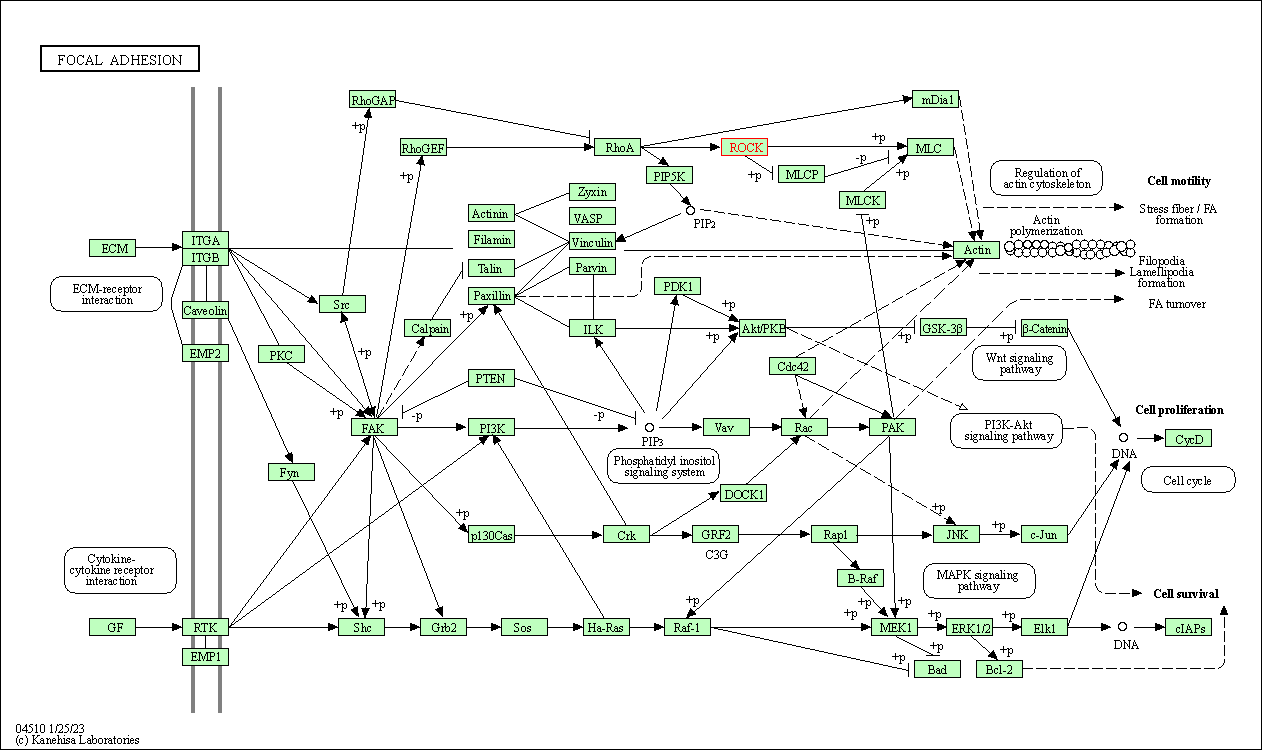
| Focal adhesion | hsa04510 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Tight junction | hsa04530 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Platelet activation | hsa04611 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Leukocyte transendothelial migration | hsa04670 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |

|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| Oxytocin signaling pathway | hsa04921 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 15 | Degree centrality | 1.61E-03 | Betweenness centrality | 4.08E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.35E-01 | Radiality | 1.41E+01 | Clustering coefficient | 3.14E-01 |
| Neighborhood connectivity | 3.75E+01 | Topological coefficient | 1.01E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Selective oral ROCK2 inhibitor down-regulates IL-21 and IL-17 secretion in human T cells via STAT3-dependent mechanism. Proc Natl Acad Sci U S A. 2014 Nov 25;111(47):16814-9. | |||||
| REF 2 | FDA Approved Drug Products from FDA Official Website. 2021. Application Number: 214783. | |||||
| REF 3 | ClinicalTrials.gov (NCT00767793) A Placebo-Controlled Study of INS117548 Ophthalmic Solution in Subjects With Glaucoma (P08650). U.S. National Institutes of Health. | |||||
| REF 4 | Novel Rho kinase inhibitors with anti-inflammatory and vasodilatory activities. J Pharmacol Exp Ther. 2007 Jan;320(1):89-98. | |||||
| REF 5 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800025959) | |||||
| REF 6 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. | |||||
| REF 7 | Rho kinase inhibitors: a patent review (2014 - 2016).Expert Opin Ther Pat. 2017 Apr;27(4):507-515. | |||||
| REF 8 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800025959) | |||||
| REF 9 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1504). | |||||
| REF 10 | Development of specific Rho-kinase inhibitors and their clinical application. Biochim Biophys Acta. 2005 Dec 30;1754(1-2):245-52. | |||||
| REF 11 | Substituted 2H-isoquinolin-1-one as potent Rho-Kinase inhibitors. Part 1: Hit-to-lead account. Bioorg Med Chem Lett. 2010 Jun 1;20(11):3235-9. | |||||
| REF 12 | Substituted 2H-isoquinolin-1-ones as potent Rho-kinase inhibitors: part 3, aryl substituted pyrrolidines. Bioorg Med Chem Lett. 2010 Jun 15;20(12):3746-9. | |||||
| REF 13 | Tetrahydroisoquinoline derivatives as highly selective and potent Rho kinase inhibitors. J Med Chem. 2010 Aug 12;53(15):5727-37. | |||||
| REF 14 | Pyridylthiazole-based ureas as inhibitors of Rho associated protein kinases (ROCK1 and 2). Medchemcomm. 2012 Jun 1;3(6):699-709. | |||||
| REF 15 | Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 Inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Antitumor Agent. J Med Chem. 2020 Feb 13;63(3):1084-1104. | |||||
| REF 16 | Identification of Selective Dual ROCK1 and ROCK2 Inhibitors Using Structure-Based Drug Design. J Med Chem. 2018 Dec 27;61(24):11074-11100. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

