Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T28713
(Former ID: TTDC00010)
|
|||||
| Target Name |
Serine/threonine-protein kinase Chk2 (RAD53)
|
|||||
| Synonyms |
hCds1; Hucds1; Checkpoint kinase 2; Cds1 homolog; Cds1; CHK2 checkpoint homolog; CHK2
Click to Show/Hide
|
|||||
| Gene Name |
CHEK2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
May also negatively regulate cell cycle progression during unperturbed cell cycles. Following activation, phosphorylates numerous effectors preferentially at the consensus sequence [L-X-R-X-X-S/T]. Regulates cell cycle checkpoint arrest through phosphorylation of CDC25A, CDC25B and CDC25C, inhibiting their activity. Inhibition of CDC25 phosphatase activity leads to increased inhibitory tyrosine phosphorylation of CDK-cyclin complexes and blocks cell cycle progression. May also phosphorylate NEK6 which is involved in G2/M cell cycle arrest. Regulates DNA repair through phosphorylation of BRCA2, enhancing the association of RAD51 with chromatin which promotes DNA repair by homologous recombination. Also stimulates the transcription of genes involved in DNA repair (including BRCA2) through the phosphorylation and activation of the transcription factor FOXM1. Regulates apoptosis through the phosphorylation of p53/TP53, MDM4 and PML. Phosphorylation of p53/TP53 at 'Ser-20' by CHEK2 may alleviate inhibition by MDM2, leading to accumulation of active p53/TP53. Phosphorylation of MDM4 may also reduce degradation of p53/TP53. Also controls the transcription of pro-apoptotic genes through phosphorylation of the transcription factor E2F1. Tumor suppressor, it may also have a DNA damage-independent function in mitotic spindle assembly by phosphorylating BRCA1. Its absence may be a cause of the chromosomal instability observed in some cancer cells. Promotes the CCAR2-SIRT1 association and is required for CCAR2-mediated SIRT1 inhibition. Serine/threonine-protein kinase which is required for checkpoint-mediated cell cycle arrest, activation of DNA repair and apoptosis in response to the presence of DNA double-strand breaks.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MSRESDVEAQQSHGSSACSQPHGSVTQSQGSSSQSQGISSSSTSTMPNSSQSSHSSSGTL
SSLETVSTQELYSIPEDQEPEDQEPEEPTPAPWARLWALQDGFANLECVNDNYWFGRDKS CEYCFDEPLLKRTDKYRTYSKKHFRIFREVGPKNSYIAYIEDHSGNGTFVNTELVGKGKR RPLNNNSEIALSLSRNKVFVFFDLTVDDQSVYPKALRDEYIMSKTLGSGACGEVKLAFER KTCKKVAIKIISKRKFAIGSAREADPALNVETEIEILKKLNHPCIIKIKNFFDAEDYYIV LELMEGGELFDKVVGNKRLKEATCKLYFYQMLLAVQYLHENGIIHRDLKPENVLLSSQEE DCLIKITDFGHSKILGETSLMRTLCGTPTYLAPEVLVSVGTAGYNRAVDCWSLGVILFIC LSGYPPFSEHRTQVSLKDQITSGKYNFIPEVWAEVSEKALDLVKKLLVVDPKARFTTEEA LRHPWLQDEDMKRKFQDLLSEENESTALPQVLAQPSTSRKRPREGEAEGAETTKRPAVCA AVL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T28EY5 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | E7850 | Drug Info | Phase 2 | Solid tumour/cancer | [2], [3] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | XL844 | Drug Info | Discontinued in Phase 1 | Solid tumour/cancer | [4] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | E7850 | Drug Info | [1] | |||
| 2 | XL844 | Drug Info | [6] | |||
| Inhibitor | [+] 8 Inhibitor drugs | + | ||||
| 1 | PMID27410995-Compound-Figure3j | Drug Info | [5] | |||
| 2 | 2-(4-Phenoxy-phenyl)-1H-benzoimidazol-5-ylamine | Drug Info | [7] | |||
| 3 | 5-Nitro-2-(4-phenoxy-phenyl)-1H-benzoimidazole | Drug Info | [7] | |||
| 4 | CCT-241533 | Drug Info | [8] | |||
| 5 | Chk2 inhibitor II | Drug Info | [9] | |||
| 6 | DEBROMOHYMENIALDISINE | Drug Info | [10] | |||
| 7 | PMID22564207C25b | Drug Info | [11] | |||
| 8 | PV-1019 | Drug Info | [12] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: adenosine diphosphate | Ligand Info | |||||
| Structure Description | Crystal structure of human Chk2 in complex with ADP | PDB:2CN5 | ||||
| Method | X-ray diffraction | Resolution | 2.25 Å | Mutation | No | [13] |
| PDB Sequence |
HMSVYPKALR
217 DEYIMSKTLG227 SGACGEVKLA237 FERKTCKKVA247 IKIISKRNVE271 TEIEILKKLN 281 HPCIIKIKNF291 FDAEDYYIVL301 ELMEGGELFD311 KVVGNKRLKE321 ATCKLYFYQM 331 LLAVQYLHEN341 GIIHRDLKPE351 NVLLSSQEED361 CLIKITDFGH371 SKILGETSLM 381 RTLCGTPTYL391 APEVLVSVGT401 AGYNRAVDCW411 SLGVILFICL421 SGYPPFSEHR 431 TQVSLKDQIT441 SGKYNFIPEV451 WAEVSEKALD461 LVKKLLVVDP471 KARFTTEEAL 481 RHPWLQDEDM491 KRKFQDLLSE501 ENE
|
|||||
|
|
LEU226
3.956
GLY227
3.457
SER228
3.664
GLY229
3.084
ALA230
3.504
CYS231
3.021
GLY232
2.673
VAL234
3.339
ALA247
3.565
LYS249
2.678
ILE286
3.737
|
|||||
| Ligand Name: CCT-241533 | Ligand Info | |||||
| Structure Description | Crystal Structure of Chk2 in complex with an inhibitor | PDB:2XBJ | ||||
| Method | X-ray diffraction | Resolution | 2.30 Å | Mutation | No | [14] |
| PDB Sequence |
VYPKALRDEY
220 IMSKTLGSGA230 GEVKLAFERK241 TCKKVAIKII251 SKALNVETEI274 EILKKLNHPC 284 IIKIKNFFDA294 EDYYIVLELM304 EGGELFDKVV314 GNKRLKEATC324 KLYFYQMLLA 334 VQYLHENGII344 HRDLKPENVL354 LSSQEEDCLI364 KITDFGHSKI374 LTSLMRTLCG 386 TPTYLAPEVL396 VSVGTAGYNR406 AVDCWSLGVI416 LFICLSGYPP426 FSEHRTQVSL 436 KDQITSGKYN446 FIPEVWAEVS456 EKALDLVKKL466 LVVDPKARFT476 TEEALRHPWL 486 QDEDMKRKFQ496 DLLSEENEST506 ALPQVL
|
|||||
|
|
LEU226
3.318
GLY227
3.669
SER228
3.880
GLY229
4.227
ALA230
4.653
GLY232
4.437
VAL234
3.785
ALA247
3.758
LYS249
4.488
ILE286
3.364
LEU301
3.560
GLU302
2.975
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
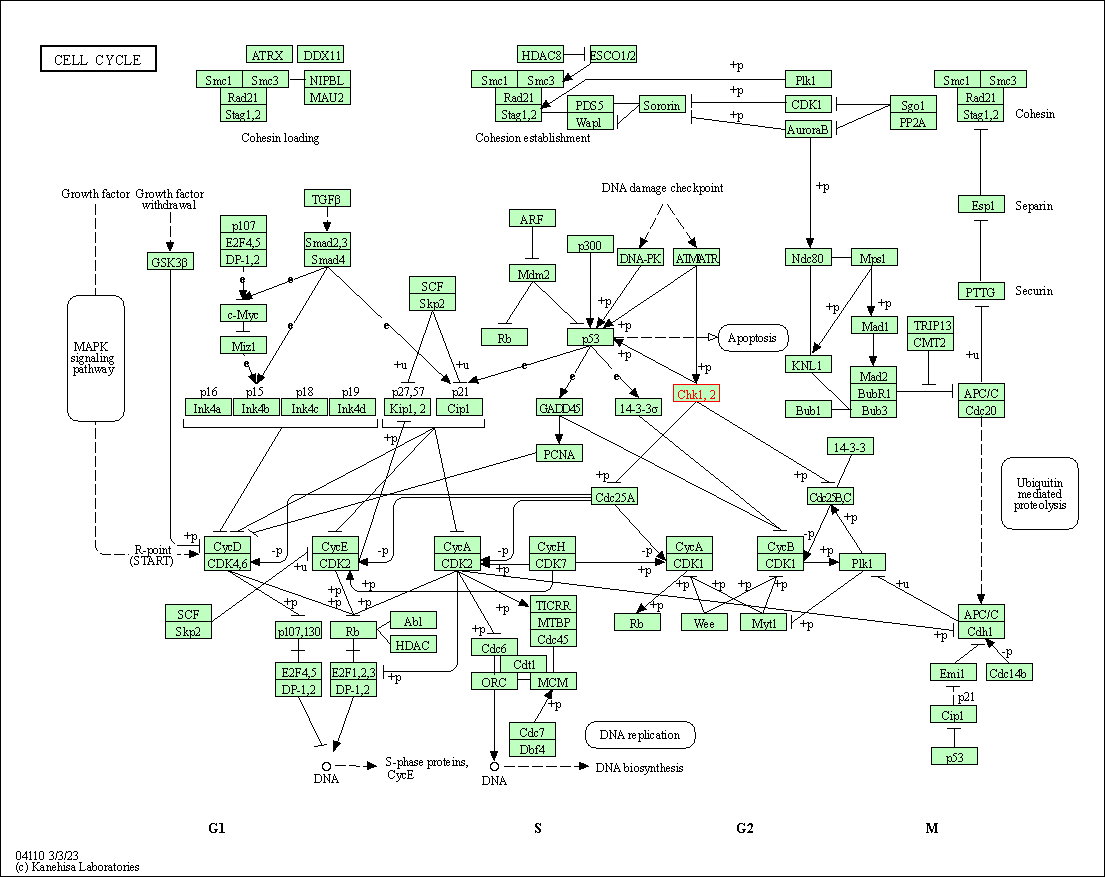
| Cell cycle | hsa04110 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
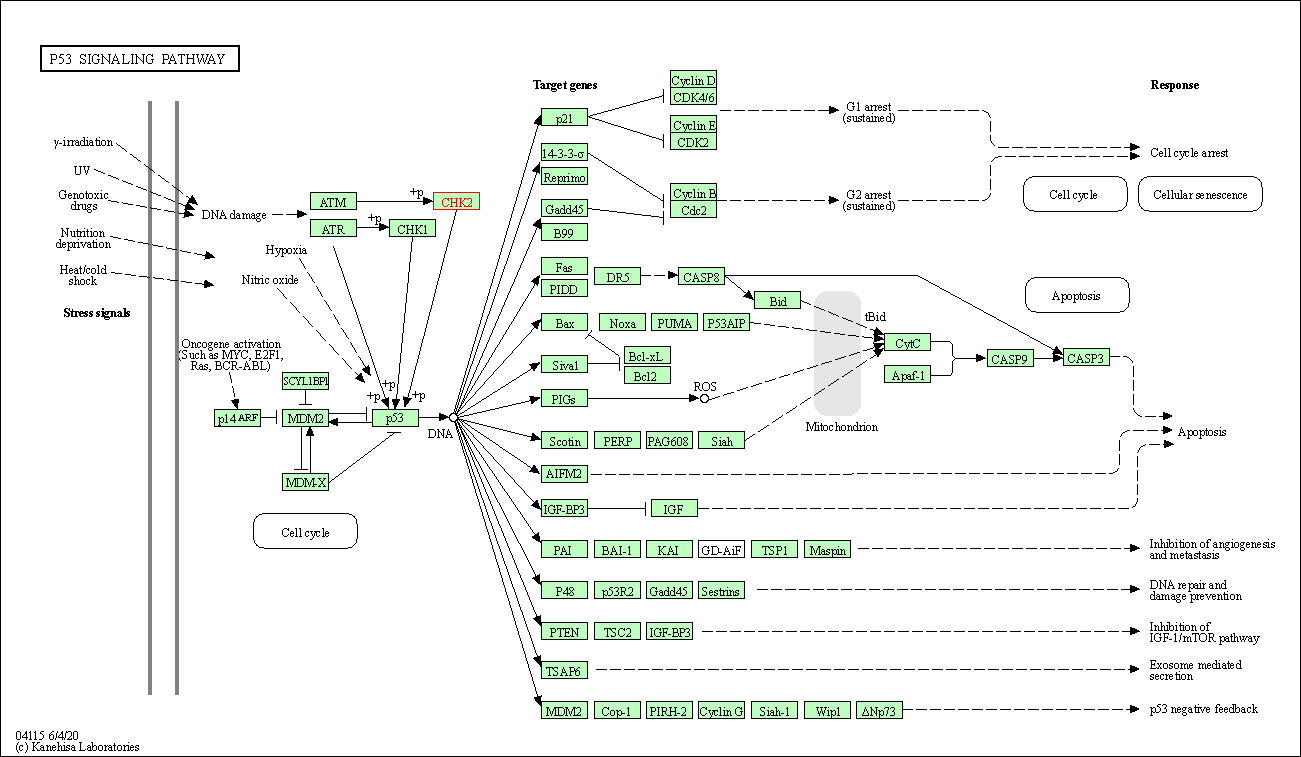
| p53 signaling pathway | hsa04115 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
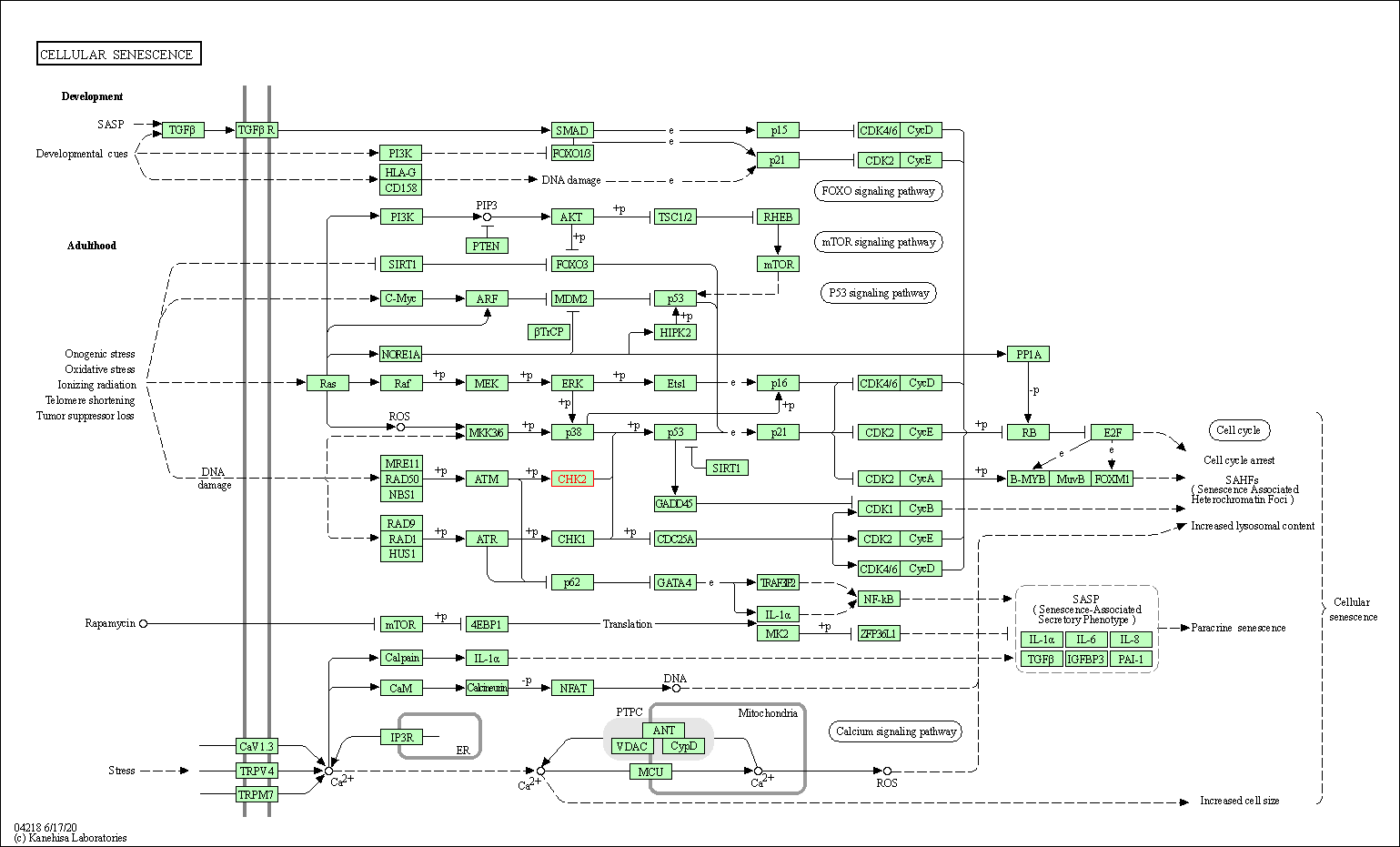
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Degree | 27 | Degree centrality | 2.90E-03 | Betweenness centrality | 2.19E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.41E-01 | Radiality | 1.42E+01 | Clustering coefficient | 2.91E-01 |
| Neighborhood connectivity | 4.76E+01 | Topological coefficient | 8.57E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Cell cycle | |||||
| 2 | p53 signaling pathway | |||||
| 3 | HTLV-I infection | |||||
| PID Pathway | [+] 4 PID Pathways | + | ||||
| 1 | ATM pathway | |||||
| 2 | FOXM1 transcription factor network | |||||
| 3 | p53 pathway | |||||
| 4 | PLK3 signaling events | |||||
| Reactome | [+] 3 Reactome Pathways | + | ||||
| 1 | Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks | |||||
| 2 | G2/M DNA damage checkpoint | |||||
| 3 | Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | |||||
| WikiPathways | [+] 8 WikiPathways | + | ||||
| 1 | DNA Damage Response | |||||
| 2 | ATM Signaling Pathway | |||||
| 3 | Integrated Pancreatic Cancer Pathway | |||||
| 4 | Prostate Cancer | |||||
| 5 | Integrated Breast Cancer Pathway | |||||
| 6 | Integrated Cancer pathway | |||||
| 7 | Cell Cycle | |||||
| 8 | miRNA Regulation of DNA Damage Response | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1988). | |||||
| REF 2 | Irofulven induces replication-dependent CHK2 activation related to p53 status. Biochem Pharmacol. 2007 Feb 15;73(4):469-80. | |||||
| REF 3 | ATM-dependent CHK2 activation induced by anticancer agent, irofulven. J Biol Chem. 2004 Sep 17;279(38):39584-92. | |||||
| REF 4 | ClinicalTrials.gov (NCT00234481) Safety Study of XL844 in Subjects With Chronic Lymphocytic Leukemia. U.S. National Institutes of Health. | |||||
| REF 5 | Ribosomal S6 kinase (RSK) modulators: a patent review.Expert Opin Ther Pat. 2016 Sep;26(9):1061-78. | |||||
| REF 6 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. | |||||
| REF 7 | Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J Med Chem. 2005 Mar 24;48(6):1873-85. | |||||
| REF 8 | CCT241533 Is a Potent and Selective Inhibitor of CHK2 that Potentiates the Cytotoxicity of PARP Inhibitors. Cancer Res. 2011 Jan 15;71(2):463-72. | |||||
| REF 9 | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20523-8. | |||||
| REF 10 | Potent inhibition of checkpoint kinase activity by a hymenialdisine-derived indoloazepine. Bioorg Med Chem Lett. 2004 Aug 16;14(16):4319-21. | |||||
| REF 11 | Discovery of an orally efficacious inhibitor of anaplastic lymphoma kinase. J Med Chem. 2012 May 24;55(10):4580-93. | |||||
| REF 12 | Cellular inhibition of checkpoint kinase 2 (Chk2) and potentiation of camptothecins and radiation by the novel Chk2 inhibitor PV1019 [7-nitro-1H-indole-2-carboxylic acid {4-[1-(guanidinohydrazone)-ethyl]-phenyl}-amide]. J Pharmacol Exp Ther. 2009 Dec;331(3):816-26. | |||||
| REF 13 | Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. EMBO J. 2006 Jul 12;25(13):3179-90. | |||||
| REF 14 | Structure-based design of potent and selective 2-(quinazolin-2-yl)phenol inhibitors of checkpoint kinase 2. J Med Chem. 2011 Jan 27;54(2):580-90. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

