Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T55398
(Former ID: TTDR00045)
|
|||||
| Target Name |
Renal carcinoma antigen NY-REN-64 (IRAK-4)
|
|||||
| Synonyms |
Interleukin-1 receptor-associated kinase 4; IRAK-4
Click to Show/Hide
|
|||||
| Gene Name |
IRAK4
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 6 Target-related Diseases | + | ||||
| 1 | Atopic eczema [ICD-11: EA80] | |||||
| 2 | Rheumatoid arthritis [ICD-11: FA20] | |||||
| 3 | Cutaneous lupus erythematosus [ICD-11: EB50-EB5Z] | |||||
| 4 | Lymphoma [ICD-11: 2A80-2A86] | |||||
| 5 | Postoperative inflammation [ICD-11: 1A00-CA43] | |||||
| 6 | Psoriasis [ICD-11: EA90] | |||||
| Function |
Serine/threonine-protein kinase that plays a critical role in initiating innate immune response against foreign pathogens. Involved in Toll-like receptor (TLR) and IL-1R signaling pathways. Is rapidly recruited by MYD88 to the receptor-signaling complex upon TLR activation to form the Myddosome together with IRAK2. Phosphorylates initially IRAK1, thus stimulating the kinase activity and intensive autophosphorylation of IRAK1. Phosphorylates E3 ubiquitin ligases Pellino proteins (PELI1, PELI2 and PELI3) to promote pellino-mediated polyubiquitination of IRAK1. Then, the ubiquitin-binding domain of IKBKG/NEMO binds to polyubiquitinated IRAK1 bringing together the IRAK1-MAP3K7/TAK1-TRAF6 complex and the NEMO-IKKA-IKKB complex. In turn, MAP3K7/TAK1 activates IKKs (CHUK/IKKA and IKBKB/IKKB) leading to NF-kappa-B nuclear translocation and activation. Alternatively, phosphorylates TIRAP to promote its ubiquitination and subsequent degradation. Phosphorylates NCF1 and regulates NADPH oxidase activation after LPS stimulation suggesting a similar mechanism during microbial infections.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MNKPITPSTYVRCLNVGLIRKLSDFIDPQEGWKKLAVAIKKPSGDDRYNQFHIRRFEALL
QTGKSPTSELLFDWGTTNCTVGDLVDLLIQNEFFAPASLLLPDAVPKTANTLPSKEAITV QQKQMPFCDKDRTLMTPVQNLEQSYMPPDSSSPENKSLEVSDTRFHSFSFYELKNVTNNF DERPISVGGNKMGEGGFGVVYKGYVNNTTVAVKKLAAMVDITTEELKQQFDQEIKVMAKC QHENLVELLGFSSDGDDLCLVYVYMPNGSLLDRLSCLDGTPPLSWHMRCKIAQGAANGIN FLHENHHIHRDIKSANILLDEAFTAKISDFGLARASEKFAQTVMTSRIVGTTAYMAPEAL RGEITPKSDIYSFGVVLLEIITGLPAVDEHREPQLLLDIKEEIEDEEKTIEDYIDKKMND ADSTSVEAMYSVASQCLHEKKNKRPDIKKVQQLLQEMTAS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T62ERY | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 6 Clinical Trial Drugs | + | ||||
| 1 | GS-5718 | Drug Info | Phase 2 | Rheumatoid arthritis | [2] | |
| 2 | Zabedosertib | Drug Info | Phase 2 | Atopic dermatitis | [3] | |
| 3 | AZD6793 | Drug Info | Phase 1 | Inflammation | [4] | |
| 4 | BAY1834845 | Drug Info | Phase 1 | Psoriasis vulgaris | [5] | |
| 5 | CA-4948 | Drug Info | Phase 1 | Lymphoma | [1] | |
| 6 | SAR444656 | Drug Info | Phase 1 | Atopic dermatitis | [6] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 34 Inhibitor drugs | + | ||||
| 1 | GS-5718 | Drug Info | [7] | |||
| 2 | Zabedosertib | Drug Info | [8] | |||
| 3 | AZD6793 | Drug Info | [9] | |||
| 4 | BAY1834845 | Drug Info | [5] | |||
| 5 | CA-4948 | Drug Info | [1] | |||
| 6 | Amidopyrazole derivative 1 | Drug Info | [10] | |||
| 7 | Amidopyrazole derivative 2 | Drug Info | [10] | |||
| 8 | Amidopyrazole derivative 3 | Drug Info | [10] | |||
| 9 | Amidopyrazole derivative 4 | Drug Info | [10] | |||
| 10 | Amidopyrazole derivative 5 | Drug Info | [10] | |||
| 11 | Amidopyrazole derivative 6 | Drug Info | [10] | |||
| 12 | Diaminopyridine analog 4 | Drug Info | [10] | |||
| 13 | Diaminopyridine analog 5 | Drug Info | [10] | |||
| 14 | Fused benzoheterocycle amide derivative 1 | Drug Info | [10] | |||
| 15 | Fused benzoheterocycle amide derivative 2 | Drug Info | [10] | |||
| 16 | Indazole amide derivative 1 | Drug Info | [10] | |||
| 17 | Indazole amide derivative 2 | Drug Info | [10] | |||
| 18 | Indazoletriazolephenyl derivative 1 | Drug Info | [10] | |||
| 19 | Indazoletriazolephenyl derivative 2 | Drug Info | [10] | |||
| 20 | Macrolactam derivative 1 | Drug Info | [10] | |||
| 21 | Macrolactam derivative 2 | Drug Info | [10] | |||
| 22 | Macrolactam derivative 3 | Drug Info | [10] | |||
| 23 | Macrolactam derivative 4 | Drug Info | [10] | |||
| 24 | Pyrazole N-1 aryl and heteroaryl derivative 1 | Drug Info | [10] | |||
| 25 | Pyrazolopyrimidine and thienopyrimidine amide derivative 1 | Drug Info | [10] | |||
| 26 | Pyrazolopyrimidine and thienopyrimidine amide derivative 2 | Drug Info | [10] | |||
| 27 | IRAK-1/4 inhibitor | Drug Info | [11] | |||
| 28 | IRAK4 inhibitor 4b | Drug Info | [12] | |||
| 29 | IRAK4 inhibitor rac-45 | Drug Info | [13] | |||
| 30 | ND-2110 | Drug Info | [14] | |||
| 31 | ND-2158 | Drug Info | [14] | |||
| 32 | PF-05313261 | Drug Info | [15] | |||
| 33 | WO2012007375C1 | Drug Info | [15] | |||
| 34 | WO2012007375C7 | Drug Info | [15] | |||
| Degrader | [+] 1 Degrader drugs | + | ||||
| 1 | SAR444656 | Drug Info | [6] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Ponatinib | Ligand Info | |||||
| Structure Description | IRAK4 in complex with Ponatinib | PDB:6EG9 | ||||
| Method | X-ray diffraction | Resolution | 2.41 Å | Mutation | No | [16] |
| PDB Sequence |
RFHSFSFYEL
173 KNVTNNFDER183 PISVGGNKMG193 EGGFGVVYKG203 YVNNTTVAVK213 KLAAITTEEL 226 KQQFDQEIKV236 MAKCQHENLV246 ELLGFSSDGD256 DLCLVYVYMP266 NGSLLDRLSC 276 LDGTPPLSWH286 MRCKIAQGAA296 NGINFLHENH306 HIHRDIKSAN316 ILLDEAFTAK 326 ISDFGVGTTA353 YMAPEALRGE363 ITPKSDIYSF373 GVVLLEIITG383 LPAVDEHREP 393 QLLLDIKEEI403 EDEEKTIEDY413 IDKKMNDADS423 TSVEAMYSVA433 SQCLHEKKNK 443 RPDIKKVQQL453 LQEMTA
|
|||||
|
|
MET192
4.422
VAL200
4.020
ALA211
3.839
LYS213
4.238
GLU233
2.743
VAL236
3.693
MET237
3.551
CYS240
4.282
LEU245
3.826
VAL246
3.186
TYR262
3.236
VAL263
3.387
|
|||||
| Ligand Name: PF-06650833 | Ligand Info | |||||
| Structure Description | Crystal structure of IRAK4 in complex with compound 30 | PDB:5UIU | ||||
| Method | X-ray diffraction | Resolution | 2.02 Å | Mutation | No | [17] |
| PDB Sequence |
FHSFSFYELK
174 NVTNNFDERP184 ISVGGNKMGE194 GGFGVVYKGY204 VNNTTVAVKK214 LAAMVDITTE 224 ELKQQFDQEI234 KVMAKCQHEN244 LVELLGFSSD254 GDDLCLVYVY264 MPNGSLLDRL 274 SCLDGTPPLS284 WHMRCKIAQG294 AANGINFLHE304 NHHIHRDIKS314 ANILLDEAFT 324 AKISDFGLAR334 ASVMRIVGTT352 AYMAPEALRG362 EITPKSDIYS372 FGVVLLEIIT 382 GLPAVDEHRE392 PQLLLDIKEE402 IEDEEKTIED412 YIDKKMNDAD422 STSVEAMYSV 432 ASQCLHEKKN442 KRPDIKKVQQ452 LLQEMT
|
|||||
|
|
MET192
3.064
GLY193
3.820
GLU194
2.737
GLY195
2.848
GLY196
4.719
PHE197
4.766
GLY198
2.708
VAL199
3.437
VAL200
2.917
ALA211
3.253
LYS213
3.112
VAL246
3.802
TYR262
2.911
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
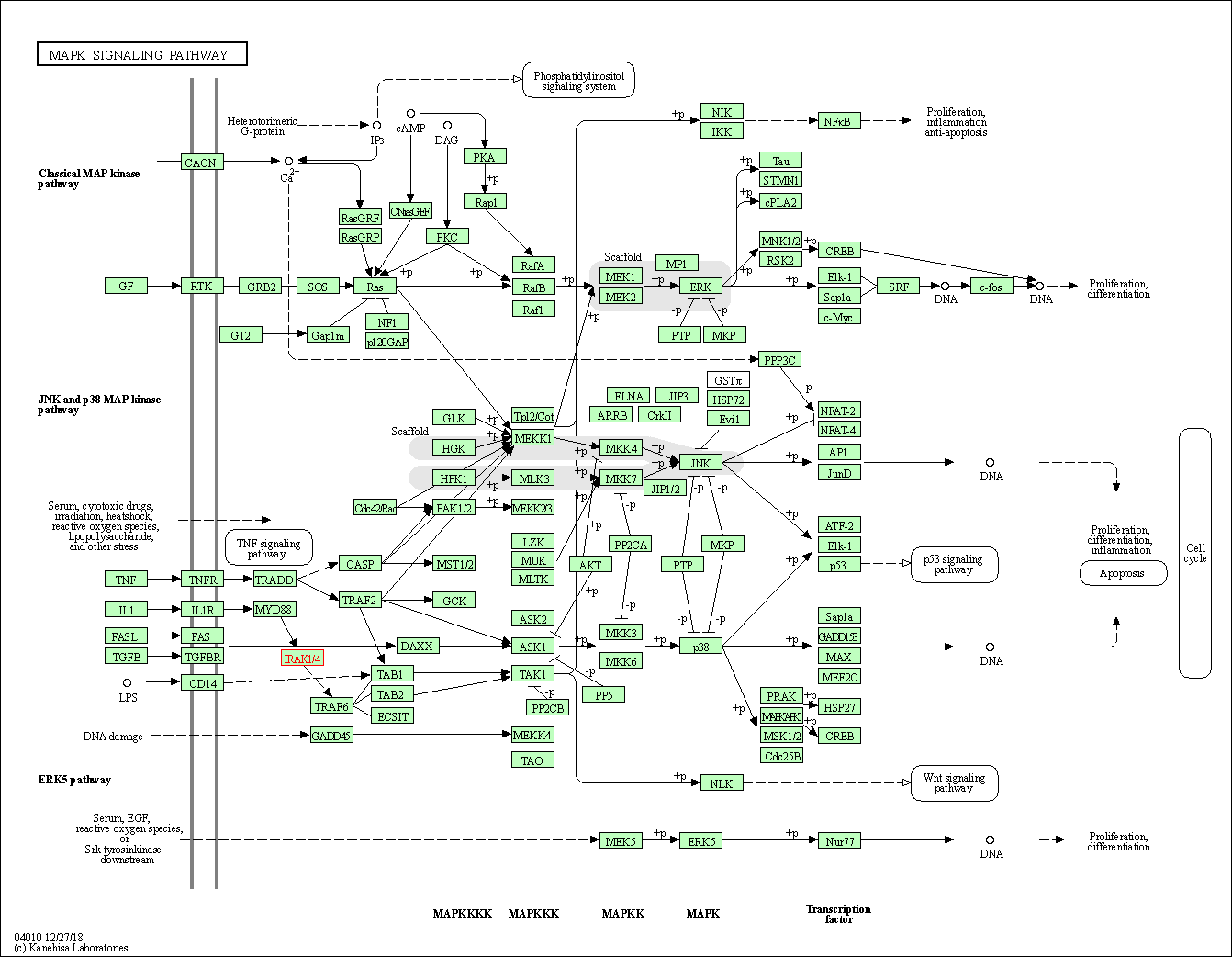
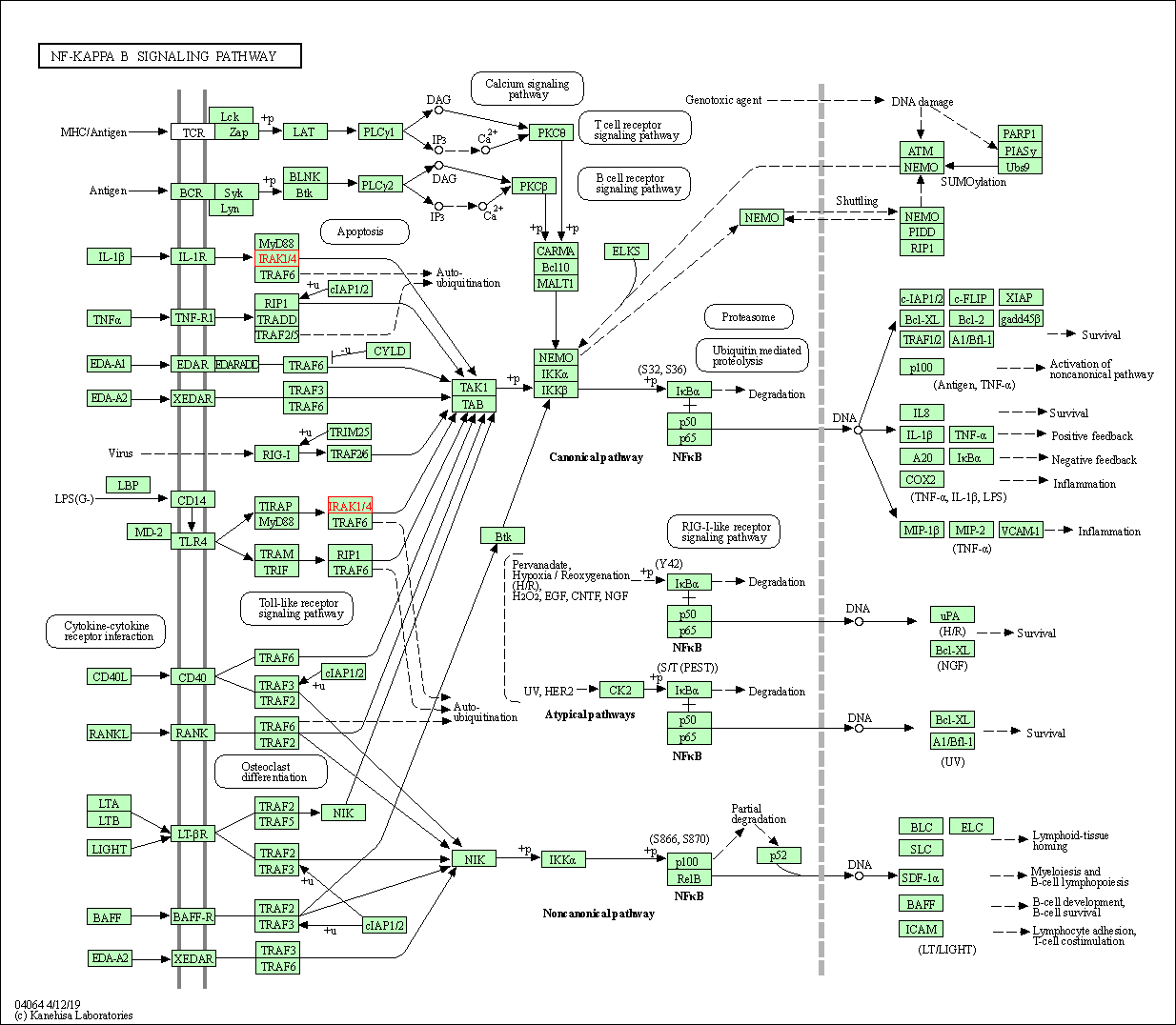
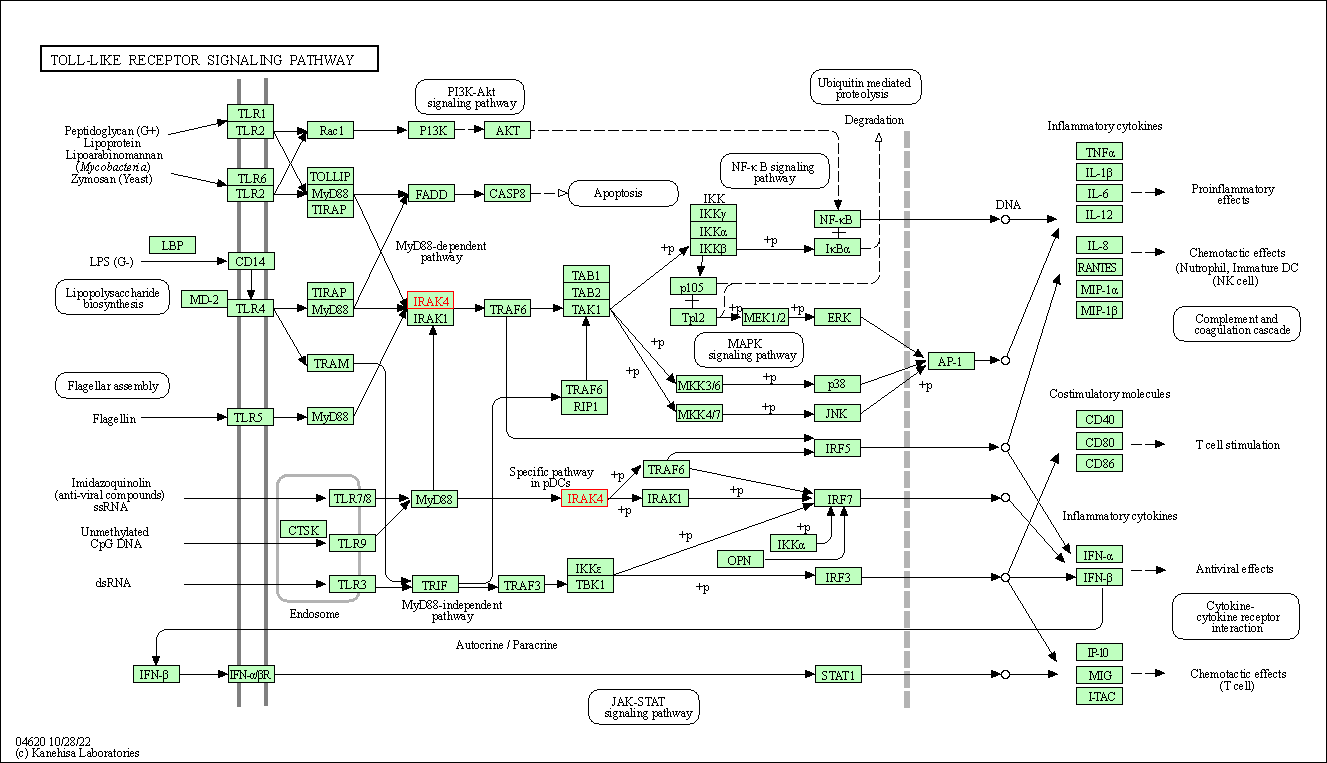
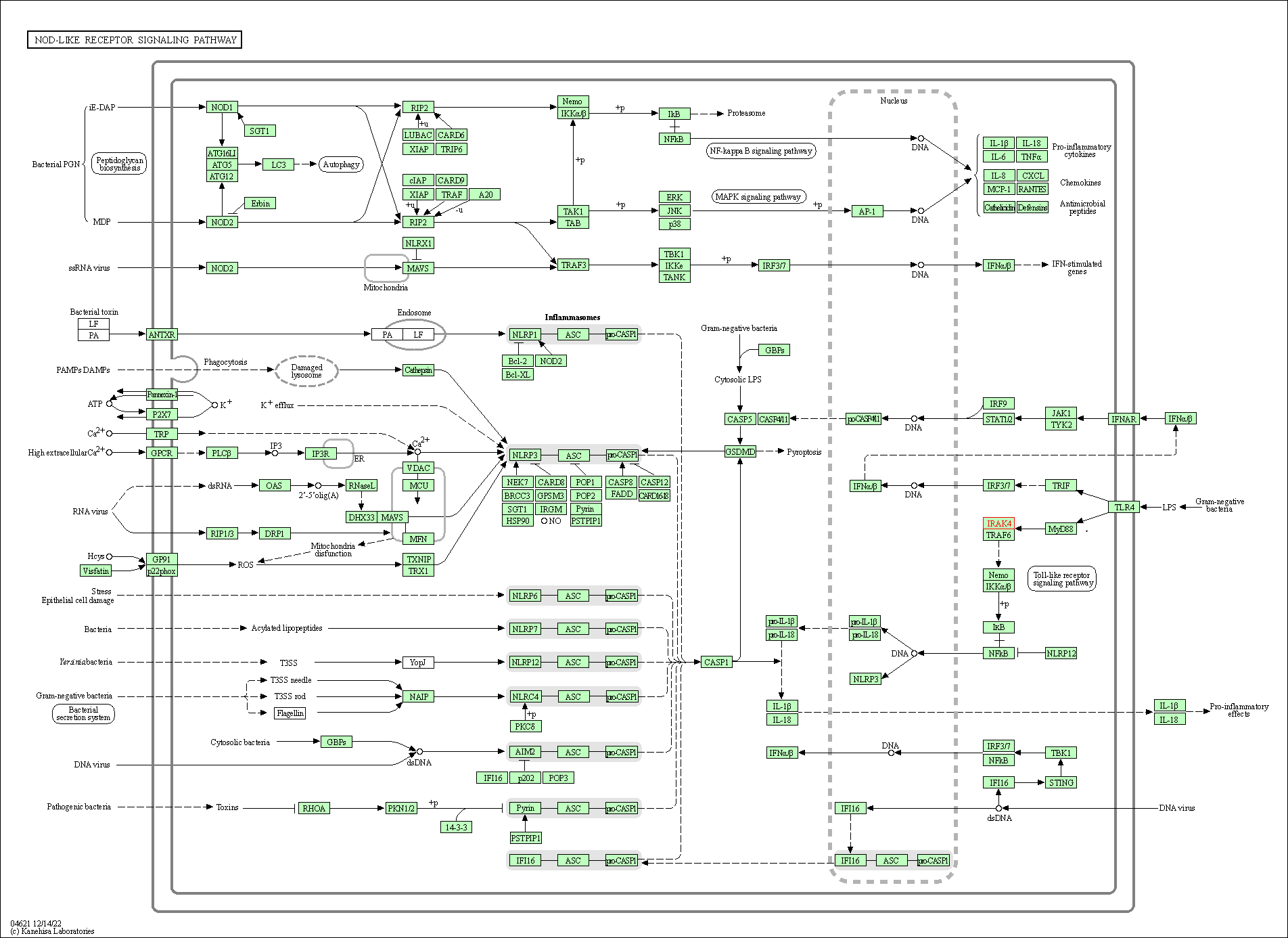
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Degree | 17 | Degree centrality | 1.83E-03 | Betweenness centrality | 3.82E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.24E-01 | Radiality | 1.39E+01 | Clustering coefficient | 5.15E-01 |
| Neighborhood connectivity | 2.92E+01 | Topological coefficient | 1.25E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | ClinicalTrials.gov (NCT05165771) A Randomized, Double-Blind, Placebo-Controlled, Multicenter, Phase 2 Proof-of-Concept Study to Evaluate Safety, Tolerability, and Efficacy of GS-5718 on Background Therapy With Conventional Synthetic Disease-modifying Antirheumatic Drug(s) (csDMARDs) in Participants With Active Rheumatoid Arthritis Who Have an Inadequate Response to Biologic DMARD(s) Treatment. U.S.National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT05656911) A Randomized, Placebo-controlled, Double-blind, Parallel-group, Multicenter Phase 2a Study to Investigate Efficacy and Safety of Zabedosertib (BAY 1834845) for the Treatment of Adult Patients With Moderate-to-severe Atopic Dermatitis. U.S.National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT05662033) A Blinded, Randomised, Placebo-controlled Study to Investigate the Safety, Tolerability, and Pharmacokinetics of an Oral Suspension of AZD6793 Following Single and Multiple Ascending Doses in Healthy Subjects, an Open-label Study to Assess the Relative Bioavailability and Food Effect of a Tablet Formulation of AZD6793 in Healthy Subjects and a Blinded, Randomised, Placebo-controlled Study to Investigate the Safety, Tolerability, and Pharmacokinetics of a Tablet Formulation of AZD6793 in Patients With Chronic Obstructive Pulmonary Disease. U.S.National Institutes of Health. | |||||
| REF 5 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 6 | Clinical pipeline report, company report or official report of Sanofi | |||||
| REF 7 | IRAK4 degrader to take on innate immunity. Nat Biotechnol. 2020 Nov;38(11):1221-1223. | |||||
| REF 8 | Oral IRAK4 inhibitor BAY-1834845 prevents acute respiratory distress syndrome. Biomed Pharmacother. 2022 Sep;153:113459. | |||||
| REF 9 | Clinical pipeline report, company report or official report of AstraZeneca | |||||
| REF 10 | Inhibitors of interleukin-1 receptor-associated kinase 4 (IRAK4): a patent review (2012-2015).Expert Opin Ther Pat. 2016 Aug;26(8):917-32. | |||||
| REF 11 | Discovery and initial SAR of inhibitors of interleukin-1 receptor-associated kinase-4. Bioorg Med Chem Lett. 2006 Jun 1;16(11):2842-5. | |||||
| REF 12 | IRAK-4 inhibitors. Part 1: a series of amides. Bioorg Med Chem Lett. 2008 Jun 1;18(11):3211-4. | |||||
| REF 13 | IRAK-4 inhibitors. Part III: a series of imidazo[1,2-a]pyridines. Bioorg Med Chem Lett. 2008 Jun 15;18(12):3656-60. | |||||
| REF 14 | Recent advances in the discovery of small molecule inhibitors of interleukin-1 receptor-associated kinase 4 (IRAK4) as a therapeutic target for inflammation and oncology disorders. J Med Chem. 2015 Jan 8;58(1):96-110. | |||||
| REF 15 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2045). | |||||
| REF 16 | Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4). J Biol Chem. 2019 Mar 22;294(12):4511-4519. | |||||
| REF 17 | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design. J Med Chem. 2017 Jul 13;60(13):5521-5542. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

