Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T86161
(Former ID: TTDR00015)
|
|||||
| Target Name |
Prolyl endopeptidase (PREP)
|
|||||
| Synonyms |
Post-proline cleaving enzyme; PREP; PE
Click to Show/Hide
|
|||||
| Gene Name |
PREP
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 4 Target-related Diseases | + | ||||
| 1 | Coeliac disease [ICD-11: DA95] | |||||
| 2 | Influenza [ICD-11: 1E30-1E32] | |||||
| 3 | Mild neurocognitive disorder [ICD-11: 6D71] | |||||
| 4 | Lung cancer [ICD-11: 2C25] | |||||
| Function |
Cleaves peptide bonds on the C-terminal side of prolyl residues within peptides that are up to approximately 30 amino acids long.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.21.26
|
|||||
| Sequence |
MLSLQYPDVYRDETAVQDYHGHKICDPYAWLEDPDSEQTKAFVEAQNKITVPFLEQCPIR
GLYKERMTELYDYPKYSCHFKKGKRYFYFYNTGLQNQRVLYVQDSLEGEARVFLDPNILS DDGTVALRGYAFSEDGEYFAYGLSASGSDWVTIKFMKVDGAKELPDVLERVKFSCMAWTH DGKGMFYNSYPQQDGKSDGTETSTNLHQKLYYHVLGTDQSEDILCAEFPDEPKWMGGAEL SDDGRYVLLSIREGCDPVNRLWYCDLQQESSGIAGILKWVKLIDNFEGEYDYVTNEGTVF TFKTNRQSPNYRVINIDFRDPEESKWKVLVPEHEKDVLEWIACVRSNFLVLCYLHDVKNI LQLHDLTTGALLKTFPLDVGSIVGYSGQKKDTEIFYQFTSFLSPGIIYHCDLTKEELEPR VFREVTVKGIDASDYQTVQIFYPSKDGTKIPMFIVHKKGIKLDGSHPAFLYGYGGFNISI TPNYSVSRLIFVRHMGGILAVANIRGGGEYGETWHKGGILANKQNCFDDFQCAAEYLIKE GYTSPKRLTINGGSNGGLLVAACANQRPDLFGCVIAQVGVMDMLKFHKYTIGHAWTTDYG CSDSKQHFEWLVKYSPLHNVKLPEADDIQYPSMLLLTADHDDRVVPLHSLKFIATLQYIV GRSRKQSNPLLIHVDTKAGHGAGKPTAKVIEEVSDMFAFIARCLNVDWIP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T16FZA | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 4 Clinical Trial Drugs | + | ||||
| 1 | ALV-003 | Drug Info | Phase 2 | Coeliac disease | [2] | |
| 2 | BAICALEIN | Drug Info | Phase 2 | Influenza virus infection | [3] | |
| 3 | ONO-1603 | Drug Info | Phase 2 | Cognitive impairment | [4] | |
| 4 | S-17092-1 | Drug Info | Phase 1 | Cognitive impairment | [5] | |
| Discontinued Drug(s) | [+] 3 Discontinued Drugs | + | ||||
| 1 | JTP-4819 | Drug Info | Discontinued in Phase 2 | Cognitive impairment | [6] | |
| 2 | Z-321 | Drug Info | Discontinued in Phase 1 | Parkinson disease | [7] | |
| 3 | BAICALIN | Drug Info | Terminated | Human immunodeficiency virus infection | [8] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 4 Modulator drugs | + | ||||
| 1 | ALV-003 | Drug Info | [9] | |||
| 2 | ONO-1603 | Drug Info | [1] | |||
| 3 | JTP-4819 | Drug Info | [13] | |||
| 4 | Z-321 | Drug Info | [14] | |||
| Inhibitor | [+] 9 Inhibitor drugs | + | ||||
| 1 | BAICALEIN | Drug Info | [10] | |||
| 2 | S-17092-1 | Drug Info | [11], [12] | |||
| 3 | BAICALIN | Drug Info | [12] | |||
| 4 | Y-29794 | Drug Info | [12] | |||
| 5 | 1-Hydroxy-1-Thio-Glycerol | Drug Info | [15] | |||
| 6 | ARI-3531 | Drug Info | [16] | |||
| 7 | Double Oxidized Cysteine | Drug Info | [15] | |||
| 8 | Monothioglycerol | Drug Info | [15] | |||
| 9 | Z-Pro-Prolinal | Drug Info | [17] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: (6s)-1-Chloro-3-[(4-Fluorobenzyl)oxy]-6-(Pyrrolidin-1-Ylcarbonyl)pyrrolo[1,2-A]pyrazin-4(6h)-One | Ligand Info | |||||
| Structure Description | Prolyl Oligopeptidase with GSK552 | PDB:3DDU | ||||
| Method | X-ray diffraction | Resolution | 1.56 Å | Mutation | Yes | [18] |
| PDB Sequence |
LSFQYPDVYR
11 DETAVQDYHG21 HKICDPYAWL31 EDPDSEQTKA41 FVEAQNKITV51 PFLEQCPIRG 61 LYKERMTELY71 DYPKYSCHFK81 KGKRYFYFYN91 TGLQNQRVLY101 VQDSLEGEAR 111 VFLDPNILSD121 DGTVALRGYA131 FSEDGEYFAY141 GLSASGSDWV151 TIKFMKVDGA 161 KELPDVLERV171 KFSCMAWTHD181 GKGMFYNSYP191 QQDGKSDGTE201 TSTNLHQKLY 211 YHVLGTDQSE221 DILCAEFPDE231 PKWMGGAELS241 DDGRYVLLSI251 REGCDPVNRL 261 WYCDLQQESS271 GIAGILKWVK281 LIDNFEGEYD291 YVTNEGTVFT301 FKTNRHSPNY 311 RVINIDFTDP321 EESKWKVLVP331 EHEKDVLEWI341 ACVRSNFLVL351 CYLHDVKNTL 361 QLHDLTTGAL371 LKIFPLDVGS381 IVGYSGQKKD391 TEIFYQFTSF401 LSPGIIYHCD 411 LTKEELEPRV421 FREVVKIDAS433 DYQTVQIFYP443 SKDGTKIPMF453 IVHKKGIKLD 463 GSHPAFLYGY473 GGFNISITPN483 YSVSRLIFVR493 HMGGILAVAN503 IRGGGEYGET 513 WHKGGILANK523 QNCFDDFQCA533 AEYLIKEGYT543 SPKRLTINGG553 SNGGLLVAAC 563 ANQRPDLFGC573 VIAQVGVMDM583 LKFHKYTIGH593 AWTTDYGCSD603 SKQHFEWLVK 613 YSPLHNVKLP623 EADDIQYPSM633 LLLTADHDDR643 VVPLHSLKFI653 ATLQYIVGRS 663 RKQNNPLLIH673 VDTKAGHGAG683 KPTAKVIEEV693 SDMFAFIARC703 LNVDWIP |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Renin-angiotensin system | hsa04614 | Affiliated Target |
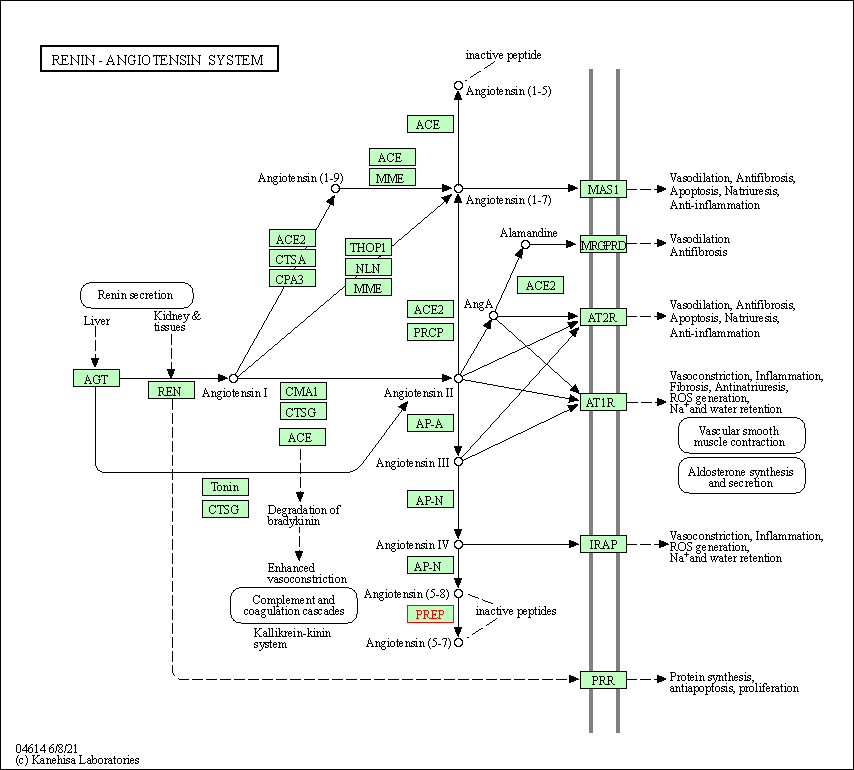
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Renin-angiotensin system | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Vasopressin synthesis | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | ONO-1603, a potential antidementia drug, shows neuroprotective effects and increases m3-muscarinic receptor mRNA levels in differentiating rat cerebellar granule neurons. Neurosci Lett. 1996 Aug 23;214(2-3):151-4. | |||||
| REF 2 | ClinicalTrials.gov (NCT00959114) Safety and Efficacy of ALV003 for the Treatment of Celiac Disease. U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT03830684) A Randomized, Double-blind, Placebo-controlled, Multicenter and Phase IIa Clinical Trial for the Effectiveness and Safety of Baicalein Tablets in the Treatment of Improve Other Aspects of Healthy Adult With Influenza Fever. U.S. National Institutes of Health. | |||||
| REF 4 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800002119) | |||||
| REF 5 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800011104) | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800006163) | |||||
| REF 7 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800003089) | |||||
| REF 8 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800007921) | |||||
| REF 9 | The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin Immunol. 2010 Mar;134(3):289-95. | |||||
| REF 10 | Baicalin, a prodrug able to reach the CNS, is a prolyl oligopeptidase inhibitor. Bioorg Med Chem. 2008 Aug 1;16(15):7516-24. | |||||
| REF 11 | Effect of S 17092, a novel prolyl endopeptidase inhibitor, on substance P and alpha-melanocyte-stimulating hormone breakdown in the rat brain. J Neurochem. 2003 Mar;84(5):919-29. | |||||
| REF 12 | Inhibitors of prolyl oligopeptidases for the therapy of human diseases: defining diseases and inhibitors. J Med Chem. 2010 May 13;53(9):3423-38. | |||||
| REF 13 | A novel prolyl endopeptidase inhibitor, JTP-4819, with potential for treating Alzheimer's disease. Behav Brain Res. 1997 Feb;83(1-2):147-51. | |||||
| REF 14 | Z-321, a prolyl endopeptidase inhibitor, augments the potentiation of synaptic transmission in rat hippocampal slices. Behav Brain Res. 1997 Feb;83(1-2):213-6. | |||||
| REF 15 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 16 | Identification of selective and potent inhibitors of fibroblast activation protein and prolyl oligopeptidase. J Med Chem. 2013 May 9;56(9):3467-77. | |||||
| REF 17 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 18 | Pyrrolidinyl pyridone and pyrazinone analogues as potent inhibitors of prolyl oligopeptidase (POP). Bioorg Med Chem Lett. 2008 Aug 1;18(15):4360-3. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

