| Drug General Information |
| Drug ID |
D06EWG
|
| Former ID |
DNC003012
|
| Drug Name |
Formic Acid
|
| Drug Type |
Small molecular drug
|
| Structure |
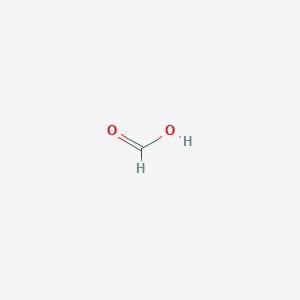
|
Download
2D MOL
3D MOL
|
| Formula |
CHO2-
|
| InChI |
InChI=1S/CH2O2/c2-1-3/h1H,(H,2,3)/p-1
|
| InChIKey |
BDAGIHXWWSANSR-UHFFFAOYSA-M
|
| CAS Number |
CAS 64-18-6
|
| PubChem Compound ID |
|
| PubChem Substance ID |
2203, 2712, 841549, 4668969, 8143755, 8150550, 10533692, 14709153, 24438659, 50073763, 50817872, 57320099, 92729992, 92741270, 103036860, 104042951, 104294385, 126406823, 126687287, 127497873, 135269870, 137002191, 142323754, 160644736, 160644737, 160644738, 163218836, 178101261, 223444138, 223444139, 223749098
|
| Target and Pathway |
| References |