Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T44458
(Former ID: TTDC00095)
|
|||||
| Target Name |
Cyclin-dependent kinase 9 (CDK9)
|
|||||
| Synonyms |
Tat-associated kinase complex catalytic subunit; TAK; Similar to cyclin-dependent kinase 9; Serine/threonine-protein kinase PITALRE; Cyclin-dependent protein kinase Cdk9; Cell division protein kinase 9; Cell division cycle 2-like protein kinase 4; CDC2L4; CDC2-related kinase; C-2K
Click to Show/Hide
|
|||||
| Gene Name |
CDK9
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 7 Target-related Diseases | + | ||||
| 1 | Acute myeloid leukaemia [ICD-11: 2A60] | |||||
| 2 | Mature B-cell lymphoma [ICD-11: 2A85] | |||||
| 3 | Lymphoma [ICD-11: 2A80-2A86] | |||||
| 4 | Malignant haematopoietic neoplasm [ICD-11: 2B33] | |||||
| 5 | Mature B-cell leukaemia [ICD-11: 2A82] | |||||
| 6 | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||||
| 7 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Member of the cyclin-dependent kinase pair (CDK9/cyclin-T) complex, also called positive transcription elongation factor b (P-TEFb), which facilitates the transition from abortive to productive elongation by phosphorylating the CTD (C-terminal domain) of the large subunit of RNA polymerase II (RNAP II) POLR2A, SUPT5H and RDBP. This complex is inactive when in the 7SK snRNP complex form. Phosphorylates EP300, MYOD1, RPB1/POLR2A and AR, and the negative elongation factors DSIF and NELF. Regulates cytokine inducible transcription networks by facilitating promoter recognition of target transcription factors (e. g. TNF-inducible RELA/p65 activation and IL-6-inducible STAT3 signaling). Promotes RNA synthesis in genetic programs for cell growth, differentiation and viral pathogenesis. P-TEFb is also involved in cotranscriptional histone modification, mRNA processing and mRNA export. Modulates a complex network of chromatin modifications including histone H2B monoubiquitination (H2Bub1), H3 lysine 4 trimethylation (H3K4me3) and H3K36me3; integrates phosphorylation during transcription with chromatin modifications to control co-transcriptional histone mRNA processing. The CDK9/cyclin-K complex has also a kinase activity towards CTD of RNAP II and can substitute for CDK9/cyclin-T P-TEFb in vitro. Replication stress response protein; the CDK9/cyclin-K complex is required for genome integrity maintenance, by promoting cell cycle recovery from replication arrest and limiting single-stranded DNA amount in response to replication stress, thus reducing the breakdown of stalled replication forks and avoiding DNA damage. In addition, probable function in DNA repair of isoform 2 via interaction with KU70/XRCC6. Promotes cardiac myocyte enlargement. RPB1/POLR2A phosphorylation on 'Ser-2' in CTD activates transcription. AR phosphorylation modulates AR transcription factor promoter selectivity and cell growth. DSIF and NELF phosphorylation promotes transcription by inhibiting their negative effect. The phosphorylation of MYOD1 enhances its transcriptional activity and thus promotes muscle differentiation. Protein kinase involved in the regulation of transcription.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.22
|
|||||
| Sequence |
MAKQYDSVECPFCDEVSKYEKLAKIGQGTFGEVFKARHRKTGQKVALKKVLMENEKEGFP
ITALREIKILQLLKHENVVNLIEICRTKASPYNRCKGSIYLVFDFCEHDLAGLLSNVLVK FTLSEIKRVMQMLLNGLYYIHRNKILHRDMKAANVLITRDGVLKLADFGLARAFSLAKNS QPNRYTNRVVTLWYRPPELLLGERDYGPPIDLWGAGCIMAEMWTRSPIMQGNTEQHQLAL ISQLCGSITPEVWPNVDNYELYEKLELVKGQKRKVKDRLKAYVRDPYALDLIDKLLVLDP AQRIDSDDALNHDFFWSDPMPSDLKGMLSTHLTSMFEYLAPPRRKGSQITQQSTNQSRNP ATTNQTEFERVF Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T32R5L | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 10 Clinical Trial Drugs | + | ||||
| 1 | Flavopiridol | Drug Info | Phase 2 | Acute myeloid leukaemia | [2], [3] | |
| 2 | P276-00 | Drug Info | Phase 2 | Mantle cell lymphoma | [4] | |
| 3 | AZD4573 | Drug Info | Phase 1 | Haematological malignancy | [3] | |
| 4 | AZD7503 | Drug Info | Phase 1 | Non-alcoholic steatohepatitis | [5] | |
| 5 | BTX-A51 | Drug Info | Phase 1 | Non-hodgkin lymphoma | [6] | |
| 6 | CYC065 | Drug Info | Phase 1 | Lymphoma | [3] | |
| 7 | RGB-286638 | Drug Info | Phase 1 | Haematological malignancy | [7], [8] | |
| 8 | SNS-032 | Drug Info | Phase 1 | Solid tumour/cancer | [9], [10] | |
| 9 | TP-1287 | Drug Info | Phase 1 | Solid tumour/cancer | [11] | |
| 10 | VIP-152 | Drug Info | Phase 1 | Non-hodgkin lymphoma | [12] | |
| Discontinued Drug(s) | [+] 3 Discontinued Drugs | + | ||||
| 1 | SCH 727965 | Drug Info | Discontinued in Phase 3 | Acute lymphoblastic leukaemia | [13], [14], [15] | |
| 2 | BAY 10-00394 | Drug Info | Discontinued in Phase 2 | Small-cell lung cancer | [16], [17] | |
| 3 | ZK 304709 | Drug Info | Discontinued in Phase 1 | Advanced solid tumour | [18] | |
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | NVP-2 | Drug Info | Preclinical | Solid tumour/cancer | [19] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 53 Inhibitor drugs | + | ||||
| 1 | Flavopiridol | Drug Info | [3] | |||
| 2 | P276-00 | Drug Info | [1] | |||
| 3 | AZD4573 | Drug Info | [3] | |||
| 4 | AZD7503 | Drug Info | [5] | |||
| 5 | BTX-A51 | Drug Info | [20] | |||
| 6 | CYC065 | Drug Info | [3] | |||
| 7 | RGB-286638 | Drug Info | [21] | |||
| 8 | SNS-032 | Drug Info | [10], [22] | |||
| 9 | TP-1287 | Drug Info | [23] | |||
| 10 | VIP-152 | Drug Info | [24] | |||
| 11 | 1,5-di-substituted pyridine derivative 1 | Drug Info | [25] | |||
| 12 | 4-(thiazol-5-yl)-pyrimidine derivative 1 | Drug Info | [25] | |||
| 13 | 4-(thiazol-5-yl)-pyrimidine derivative 2 | Drug Info | [25] | |||
| 14 | 5-fluoro-N-(pyridin-2-yl)pyridin-2-amine derivative 1 | Drug Info | [25] | |||
| 15 | Alkyl sulfone derivative 1 | Drug Info | [25] | |||
| 16 | Aminoarylpyridine derivative 1 | Drug Info | [25] | |||
| 17 | Aryl pyrimidine derivative 1 | Drug Info | [25] | |||
| 18 | Benzothiazine derivative 1 | Drug Info | [25] | |||
| 19 | Bipyridine derivative 1 | Drug Info | [25] | |||
| 20 | Diaryl amine derivative 1 | Drug Info | [25] | |||
| 21 | Flavopiridol analog 1 | Drug Info | [25] | |||
| 22 | Indole-based analog 13 | Drug Info | [25] | |||
| 23 | N-(pyridin-2-yl)pyridine methylsulfone derivative 1 | Drug Info | [25] | |||
| 24 | N-(pyridin-2-yl)pyrimidin-4-amine derivative 1 | Drug Info | [25] | |||
| 25 | N-(pyridin-2-yl)pyrimidin-4-amine derivative 2 | Drug Info | [25] | |||
| 26 | N-phenyl-pyrimidin-4-amine derivative 1 | Drug Info | [25] | |||
| 27 | Nitrogen mustard derivative 1 | Drug Info | [25] | |||
| 28 | Oxazolyl methylthiothiazole derivative 1 | Drug Info | [25] | |||
| 29 | Phenylpyridine derivative 1 | Drug Info | [25] | |||
| 30 | Phenylpyridine derivative 2 | Drug Info | [25] | |||
| 31 | PMID26161698-Compound-25 | Drug Info | [25] | |||
| 32 | PMID26161698-Compound-32 | Drug Info | [25] | |||
| 33 | Pyrazinylpyridine derivative 1 | Drug Info | [25] | |||
| 34 | Pyrazolo[1,5-a]-1,3,5-triazine derivative 1 | Drug Info | [25] | |||
| 35 | Roscovitine derivative 1 | Drug Info | [25] | |||
| 36 | Tricyclic benzimidazole derivative 1 | Drug Info | [25] | |||
| 37 | SCH 727965 | Drug Info | [1] | |||
| 38 | BAY 10-00394 | Drug Info | [26] | |||
| 39 | ZK 304709 | Drug Info | [1] | |||
| 40 | NVP-2 | Drug Info | [27] | |||
| 41 | 3-((3,5-diamino-1H-pyrazol-4-yl)diazenyl)phenol | Drug Info | [28] | |||
| 42 | 4-(phenyldiazenyl)-1H-pyrazole-3,5-diamine | Drug Info | [28] | |||
| 43 | 4-[(3,5-diamino-1H-pyrazol-4-yl)diazenyl]phenol | Drug Info | [28] | |||
| 44 | MERIOLIN 1 | Drug Info | [30] | |||
| 45 | MERIOLIN 2 | Drug Info | [30] | |||
| 46 | MERIOLIN 3 | Drug Info | [30] | |||
| 47 | MERIOLIN 4 | Drug Info | [30] | |||
| 48 | MERIOLIN 5 | Drug Info | [30] | |||
| 49 | MERIOLIN 6 | Drug Info | [30] | |||
| 50 | MERIOLIN 7 | Drug Info | [30] | |||
| 51 | MERIOLIN 8 | Drug Info | [30] | |||
| 52 | PMID19115845C89S | Drug Info | [31] | |||
| 53 | PMID20873740C18 | Drug Info | [32] | |||
| Binder | [+] 1 Binder drugs | + | ||||
| 1 | Deschloroflavopiridol | Drug Info | [29] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenosine monophosphate | Ligand Info | |||||
| Structure Description | The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat | PDB:4IMY | ||||
| Method | X-ray diffraction | Resolution | 2.94 Å | Mutation | No | [33] |
| PDB Sequence |
VECPFCDEVS
17 KYEKLAKIGQ27 GTFGEVFKAR37 HRKTGQKVAL47 KKVLMENEKE57 GFPITALREI 67 KILQLLKHEN77 VVNLIEICRT87 KGSIYLVFDF105 CEHDLAGLLS115 NVLVKFTLSE 125 IKRVMQMLLN135 GLYYIHRNKI145 LHRDMKAANV155 LITRDGVLKL165 ADFGLARAFS 175 LAKNSQPNRY185 NRVVTLWYRP196 PELLLGERDY206 GPPIDLWGAG216 CIMAEMWTRS 226 PIMQGNTEQH236 QLALISQLCG246 SITPEVWPNV256 DNYELYEKLE266 LVKGQKRKVK 276 DRLKAYVRDP286 YALDLIDKLL296 VLDPAQRIDS306 DDALNHDFFW316 SDPMPSDLKG 326 MLST
|
|||||
|
|
ILE25
2.705
VAL33
3.512
ALA46
2.909
LYS48
1.936
GLU66
2.514
LEU70
2.457
VAL79
2.433
ASN80
4.828
LEU101
4.298
PHE103
2.152
ASP104
2.184
|
|||||
| Ligand Name: Adenosine | Ligand Info | |||||
| Structure Description | crystal structure of P-TEFb complex with AFF4 and Tat | PDB:4OGR | ||||
| Method | X-ray diffraction | Resolution | 3.00 Å | Mutation | No | [34] |
| PDB Sequence |
VECPFCDEVS
17 KYEKLAKIGQ27 GTFGEVFKAR37 HRKTGQKVAL47 KKVLMENEKE57 GFPITALREI 67 KILQLLKHEN77 VVNLIEICRT87 KGSIYLVFDF105 CEHDLAGLLS115 NVLVKFTLSE 125 IKRVMQMLLN135 GLYYIHRNKI145 LHRDMKAANV155 LITRDGVLKL165 ADFGLARAFS 175 LAKNSQPNRY185 NRVVTLWYRP196 PELLLGERDY206 GPPIDLWGAG216 CIMAEMWTRS 226 PIMQGNTEQH236 QLALISQLCG246 SITPEVWPNV256 DNYELYEKLE266 LVKGQKRKVK 276 DRLKAYVRDP286 YALDLIDKLL296 VLDPAQRIDS306 DDALNHDFFW316 SDPMPSDLKG 326 MLST
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
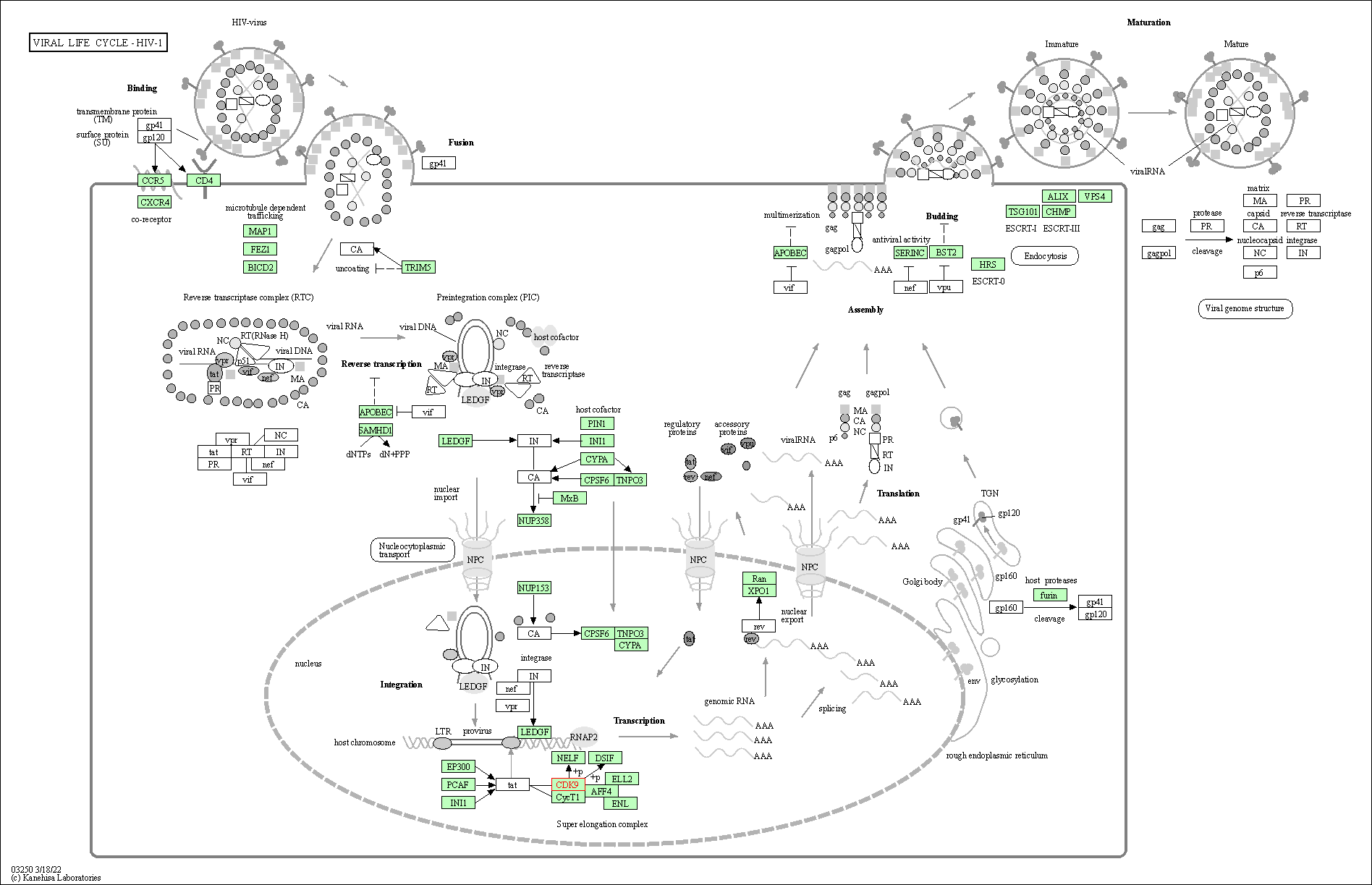
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Viral life cycle - HIV-1 | hsa03250 | Affiliated Target |

|
| Class: Genetic Information Processing => Information processing in viruses | Pathway Hierarchy | ||
| Degree | 25 | Degree centrality | 2.69E-03 | Betweenness centrality | 2.34E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.49E-01 | Radiality | 1.43E+01 | Clustering coefficient | 1.67E-01 |
| Neighborhood connectivity | 4.56E+01 | Topological coefficient | 6.76E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Transcriptional misregulation in cancer | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | EGFR1 Signaling Pathway | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription | |||||
| WikiPathways | [+] 7 WikiPathways | + | ||||
| 1 | Cardiac Hypertrophic Response | |||||
| 2 | Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer | |||||
| 3 | Host Interactions of HIV factors | |||||
| 4 | HIV Life Cycle | |||||
| 5 | IL-9 Signaling Pathway | |||||
| 6 | RNA Polymerase II Transcription | |||||
| 7 | MicroRNAs in cardiomyocyte hypertrophy | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009 Jul;8(7):547-66. | |||||
| REF 2 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 3 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 4 | A phase II, single-arm, open-label, multicenter study to evaluate the efficacy and safety of P276-00, a cyclin-dependent kinase inhibitor, in patients with relapsed or refractory mantle cell lymphoma. Clin Lymphoma Myeloma Leuk. 2015 Jul;15(7):392-7. | |||||
| REF 5 | ClinicalTrials.gov (NCT05560607) An Open-label, Non-randomized, Multiple-dose Study to Assess the Knockdown of Hepatic HSD17B13 mRNA Expression, Pharmacokinetics, Safety, and Tolerability Following Administration of AZD7503 in Participants With Non-alcoholic Fatty Liver Disease. U.S.National Institutes of Health. | |||||
| REF 6 | ClinicalTrials.gov (NCT04872166) A Study of BTX-A51 in People With Advanced Solid Tumor or Non-Hodgkin Lymphoma. U.S. National Institutes of Health. | |||||
| REF 7 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7744). | |||||
| REF 8 | ClinicalTrials.gov (NCT01168882) Safety and Tolerability of RGB-286638 in Patients With Selected, Relapsed or Refractory Hematological Malignancies. U.S. National Institutes of Health. | |||||
| REF 9 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5670). | |||||
| REF 10 | Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood. 2009 May 7;113(19):4637-45. | |||||
| REF 11 | ClinicalTrials.gov (NCT03604783) Phase 1, First-in-human Study of Oral TP-1287 in Patients With Advanced Solid Tumors. U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT04978779) An Open-label, Multicenter Phase 1/1b Study to Characterize Safety, Tolerability, Preliminary Antitumor Activity, Pharmacokinetics, and Pharmacodynamics of VIP152 Monotherapy or Combination Therapy in Subjects With High-risk Chronic Lymphocytic Leukemia or Richter Syndrome. U.S.National Institutes of Health. | |||||
| REF 13 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7379). | |||||
| REF 14 | Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood. 2015 Jan 15;125(3):443-8. | |||||
| REF 15 | Cyclin-dependent kinase inhibitor Dinaciclib (SCH727965) inhibits pancreatic cancer growth and progression in murine xenograft models. Cancer Biol Ther. 2011 Oct 1;12(7):598-609. | |||||
| REF 16 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7874). | |||||
| REF 17 | BAY 1000394, a novel cyclin-dependent kinase inhibitor, with potent antitumor activity in mono- and in combination treatment upon oral application. Mol Cancer Ther. 2012 Oct;11(10):2265-73. | |||||
| REF 18 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800022386) | |||||
| REF 19 | Kinase inhibitors: the road ahead. Nat Rev Drug Discov. 2018 May;17(5):353-377. | |||||
| REF 20 | Clinical pipeline report, company report or official report of BioTheryX. | |||||
| REF 21 | Small-molecule multi-targeted kinase inhibitor RGB-286638 triggers P53-dependent and -independent anti-multiple myeloma activity through inhibition of transcriptional CDKs. Leukemia. 2013 Dec;27(12):2366-75. | |||||
| REF 22 | Development of cell-cycle inhibitors for cancer therapy. Curr Oncol. 2009 Mar;16(2):36-43. | |||||
| REF 23 | Clinical pipeline report, company report or official report of Sumitomo Dainippon Pharma. | |||||
| REF 24 | VIP152 is a selective CDK9 inhibitor with pre-clinical in vitro and in vivo efficacy in chronic lymphocytic leukemia. Leukemia. 2023 Feb;37(2):326-338. | |||||
| REF 25 | Cyclin-dependent kinase inhibitors for cancer therapy: a patent review (2009 - 2014).Expert Opin Ther Pat. 2015;25(9):953-70. | |||||
| REF 26 | National Cancer Institute Drug Dictionary (drug id 770319). | |||||
| REF 27 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1981). | |||||
| REF 28 | 4-arylazo-3,5-diamino-1H-pyrazole CDK inhibitors: SAR study, crystal structure in complex with CDK2, selectivity, and cellular effects. J Med Chem. 2006 Nov 2;49(22):6500-9. | |||||
| REF 29 | Pharmacological inhibitors of cyclin-dependent kinases. Trends Pharmacol Sci. 2002 Sep;23(9):417-25. | |||||
| REF 30 | Meriolins (3-(pyrimidin-4-yl)-7-azaindoles): synthesis, kinase inhibitory activity, cellular effects, and structure of a CDK2/cyclin A/meriolin com... J Med Chem. 2008 Feb 28;51(4):737-51. | |||||
| REF 31 | First Cdc7 kinase inhibitors: pyrrolopyridinones as potent and orally active antitumor agents. 2. Lead discovery. J Med Chem. 2009 Jan 22;52(2):293-307. | |||||
| REF 32 | Cdc7 kinase inhibitors: 5-heteroaryl-3-carboxamido-2-aryl pyrroles as potential antitumor agents. 1. Lead finding. J Med Chem. 2010 Oct 28;53(20):7296-315. | |||||
| REF 33 | The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat. Elife. 2013 Mar 5;2:e00327. | |||||
| REF 34 | AFF4 binding to Tat-P-TEFb indirectly stimulates TAR recognition of super elongation complexes at the HIV promoter. Elife. 2014 Apr 24;3:e02375. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

