Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T16769
(Former ID: TTDI02333)
|
|||||
| Target Name |
X-linked inhibitor of apoptosis protein (XIAP)
|
|||||
| Synonyms |
hILP; hIAP3; hIAP-3; Xlinked IAP; X-linked IAP; RING-type E3 ubiquitin transferase XIAP; Inhibitor of apoptosis protein 3; ILP; IAPlike protein; IAP3; IAP-like protein; IAP-3; E3 ubiquitinprotein ligase XIAP; E3 ubiquitin-protein ligase XIAP; Baculoviral IAP repeatcontaining protein 4; Baculoviral IAP repeat-containing protein 4; BIRC4; API3
Click to Show/Hide
|
|||||
| Gene Name |
XIAP
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Metastatic lymph node neoplasm [ICD-11: 2D60] | |||||
| Function |
Acts as a direct caspase inhibitor. Directly bind to the active site pocket of CASP3 and CASP7 and obstructs substrate entry. Inactivates CASP9 by keeping it in a monomeric, inactive state. Acts as an E3 ubiquitin-protein ligase regulating NF-kappa-B signaling and the target proteins for its E3 ubiquitin-protein ligase activity include: RIPK1, CASP3, CASP7, CASP8, CASP9, MAP3K2/MEKK2, DIABLO/SMAC, AIFM1, CCS and BIRC5/survivin. Ubiquitinion of CCS leads to enhancement of its chaperone activity toward its physiologic target, SOD1, rather than proteasomal degradation. Ubiquitinion of MAP3K2/MEKK2 and AIFM1 does not lead to proteasomal degradation. Plays a role in copper homeostasis by ubiquitinationg COMMD1 and promoting its proteasomal degradation. Can also function as E3 ubiquitin-protein ligase of the NEDD8 conjugation pathway, targeting effector caspases for neddylation and inactivation. Regulates the BMP signaling pathway and the SMAD and MAP3K7/TAK1 dependent pathways leading to NF-kappa-B and JNK activation. Acts as an important regulator of innate immune signaling via regulation of Nodlike receptors (NLRs). Protects cells from spontaneous formation of the ripoptosome, a large multi-protein complex that has the capability to kill cancer cells in a caspase-dependent and caspase-independent manner. Suppresses ripoptosome formation by ubiquitinating RIPK1 and CASP8. Acts as a positive regulator of Wnt signaling and ubiquitinates TLE1, TLE2, TLE3, TLE4 and AES. Ubiquitination of TLE3 results in inhibition of its interaction with TCF7L2/TCF4 thereby allowing efficient recruitment and binding of the transcriptional coactivator beta-catenin to TCF7L2/TCF4 that is required to initiate a Wnt-specific transcriptional program. Multi-functional protein which regulates not only caspases and apoptosis, but also modulates inflammatory signaling and immunity, copper homeostasis, mitogenic kinase signaling, cell proliferation, as well as cell invasion and metastasis.
Click to Show/Hide
|
|||||
| BioChemical Class |
Acyltransferase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.3.2.27
|
|||||
| Sequence |
MTFNSFEGSKTCVPADINKEEEFVEEFNRLKTFANFPSGSPVSASTLARAGFLYTGEGDT
VRCFSCHAAVDRWQYGDSAVGRHRKVSPNCRFINGFYLENSATQSTNSGIQNGQYKVENY LGSRDHFALDRPSETHADYLLRTGQVVDISDTIYPRNPAMYSEEARLKSFQNWPDYAHLT PRELASAGLYYTGIGDQVQCFCCGGKLKNWEPCDRAWSEHRRHFPNCFFVLGRNLNIRSE SDAVSSDRNFPNSTNLPRNPSMADYEARIFTFGTWIYSVNKEQLARAGFYALGEGDKVKC FHCGGGLTDWKPSEDPWEQHAKWYPGCKYLLEQKGQEYINNIHLTHSLEECLVRTTEKTP SLTRRIDDTIFQNPMVQEAIRMGFSFKDIKKIMEEKIQISGSNYKSLEVLVADLVNAQKD SMQDESSQTSLQKEISTEEQLRRLQEEKLCKICMDRNIAIVFVPCGHLVTCKQCAEAVDK CPMCYTVITFKQKIFMS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T14LSK | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 6 Clinical Trial Drugs | + | ||||
| 1 | AT-406 | Drug Info | Phase 3 | Squamous head and neck cell carcinom | [2] | |
| 2 | Birinapant | Drug Info | Phase 2 | Lymphoma | [3], [4] | |
| 3 | Phenoxodiol | Drug Info | Phase 1/2 | Ovarian cancer | [5] | |
| 4 | APG-1387 | Drug Info | Phase 1 | Haematological malignancy | [6] | |
| 5 | GDC-0152 | Drug Info | Phase 1 | Obesity | [7] | |
| 6 | HGS-1029 | Drug Info | Phase 1 | Haematological malignancy | [8] | |
| Mode of Action | [+] 3 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | AT-406 | Drug Info | [9], [10] | |||
| Modulator | [+] 3 Modulator drugs | + | ||||
| 1 | Birinapant | Drug Info | [1], [11] | |||
| 2 | Phenoxodiol | Drug Info | [12] | |||
| 3 | GDC-0152 | Drug Info | [7] | |||
| Antagonist | [+] 36 Antagonist drugs | + | ||||
| 1 | APG-1387 | Drug Info | [13] | |||
| 2 | HGS-1029 | Drug Info | [14] | |||
| 3 | PMID25980951-Compound-1 | Drug Info | [15] | |||
| 4 | PMID25980951-Compound-10 | Drug Info | [15] | |||
| 5 | PMID25980951-Compound-11 | Drug Info | [15] | |||
| 6 | PMID25980951-Compound-12 | Drug Info | [15] | |||
| 7 | PMID25980951-Compound-13 | Drug Info | [15] | |||
| 8 | PMID25980951-Compound-14 | Drug Info | [15] | |||
| 9 | PMID25980951-Compound-15 | Drug Info | [15] | |||
| 10 | PMID25980951-Compound-19 | Drug Info | [15] | |||
| 11 | PMID25980951-Compound-20 | Drug Info | [15] | |||
| 12 | PMID25980951-Compound-21 | Drug Info | [15] | |||
| 13 | PMID25980951-Compound-22 | Drug Info | [15] | |||
| 14 | PMID25980951-Compound-26 | Drug Info | [15] | |||
| 15 | PMID25980951-Compound-27 | Drug Info | [15] | |||
| 16 | PMID25980951-Compound-28 | Drug Info | [15] | |||
| 17 | PMID25980951-Compound-29 | Drug Info | [15] | |||
| 18 | PMID25980951-Compound-3 | Drug Info | [15] | |||
| 19 | PMID25980951-Compound-30 | Drug Info | [15] | |||
| 20 | PMID25980951-Compound-31 | Drug Info | [15] | |||
| 21 | PMID25980951-Compound-32 | Drug Info | [15] | |||
| 22 | PMID25980951-Compound-33 | Drug Info | [15] | |||
| 23 | PMID25980951-Compound-36 | Drug Info | [15] | |||
| 24 | PMID25980951-Compound-37 | Drug Info | [15] | |||
| 25 | PMID25980951-Compound-38 | Drug Info | [15] | |||
| 26 | PMID25980951-Compound-39 | Drug Info | [15] | |||
| 27 | PMID25980951-Compound-4 | Drug Info | [15] | |||
| 28 | PMID25980951-Compound-40 | Drug Info | [15] | |||
| 29 | PMID25980951-Compound-41 | Drug Info | [15] | |||
| 30 | PMID25980951-Compound-42 | Drug Info | [15] | |||
| 31 | PMID25980951-Compound-43 | Drug Info | [15] | |||
| 32 | PMID25980951-Compound-44 | Drug Info | [15] | |||
| 33 | PMID25980951-Compound-45 | Drug Info | [15] | |||
| 34 | PMID25980951-Compound-5 | Drug Info | [15] | |||
| 35 | PMID25980951-Compound-6 | Drug Info | [15] | |||
| 36 | PMID25980951-Compound-7 | Drug Info | [15] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: PMID25980951-Compound-16 | Ligand Info | |||||
| Structure Description | Crystal structure of XIAP BIR3 with T3256336 | PDB:4HY0 | ||||
| Method | X-ray diffraction | Resolution | 2.84 Å | Mutation | No | [16] |
| PDB Sequence |
TNLPRNPSMA
263 DYEARIFTFG273 TWIYSVNKEQ283 LARAGFYALG293 EGDKVKCFHC303 GGGLTDWKPS 313 EDPWEQHAKW323 YPGCKYLLEQ333 KGQEYINNIH343 LTHSLE
|
|||||
|
|
||||||
| Ligand Name: (3~{S},6~{S},7~{S},9~{a}~{S})-~{N}-[(4-~{tert}-butylphenyl)methyl]-7-(hydroxymethyl)-6-[[(2~{S})-2-(methylamino)butanoyl]amino]-5-oxidanylidene-1,2,3,6,7,8,9,9~{a}-octahydropyrrolo[1,2-a]azepine-3-carboxamide | Ligand Info | |||||
| Structure Description | Crystal structure of XIAP-BIR3 in complex with a cIAP1-selective SM | PDB:6EY2 | ||||
| Method | X-ray diffraction | Resolution | 2.70 Å | Mutation | No | [17] |
| PDB Sequence |
TNLPRNPSMA
263 DYEARIFTFG273 TWIYSVNKEQ283 LARAGFYALG293 EGDKVKCFHC303 GGGLTDWKPS 313 EDPWEQHAKW323 YPGCKYLLEQ333 KGQEYINNIH343 LTHSLEECL
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
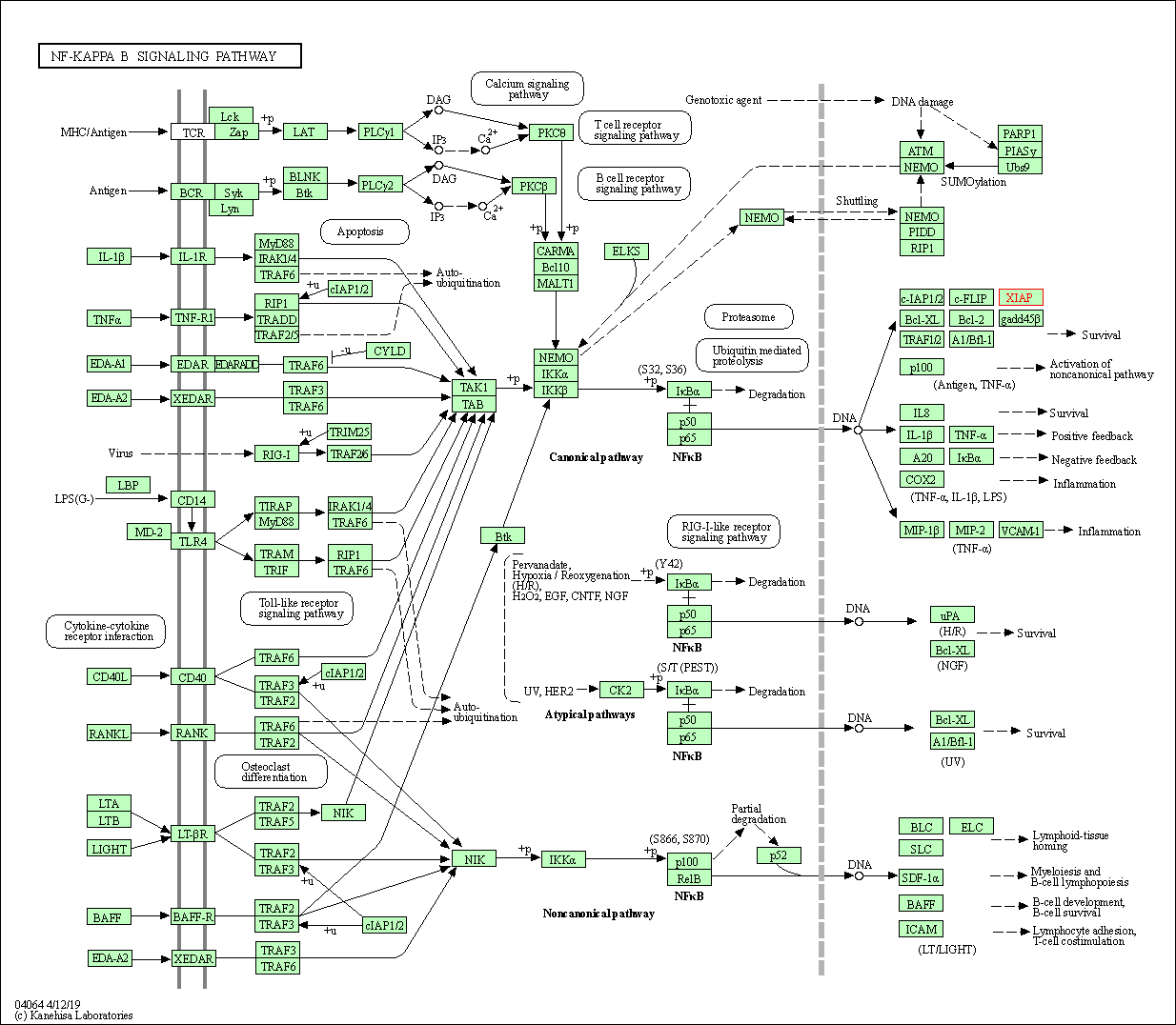
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
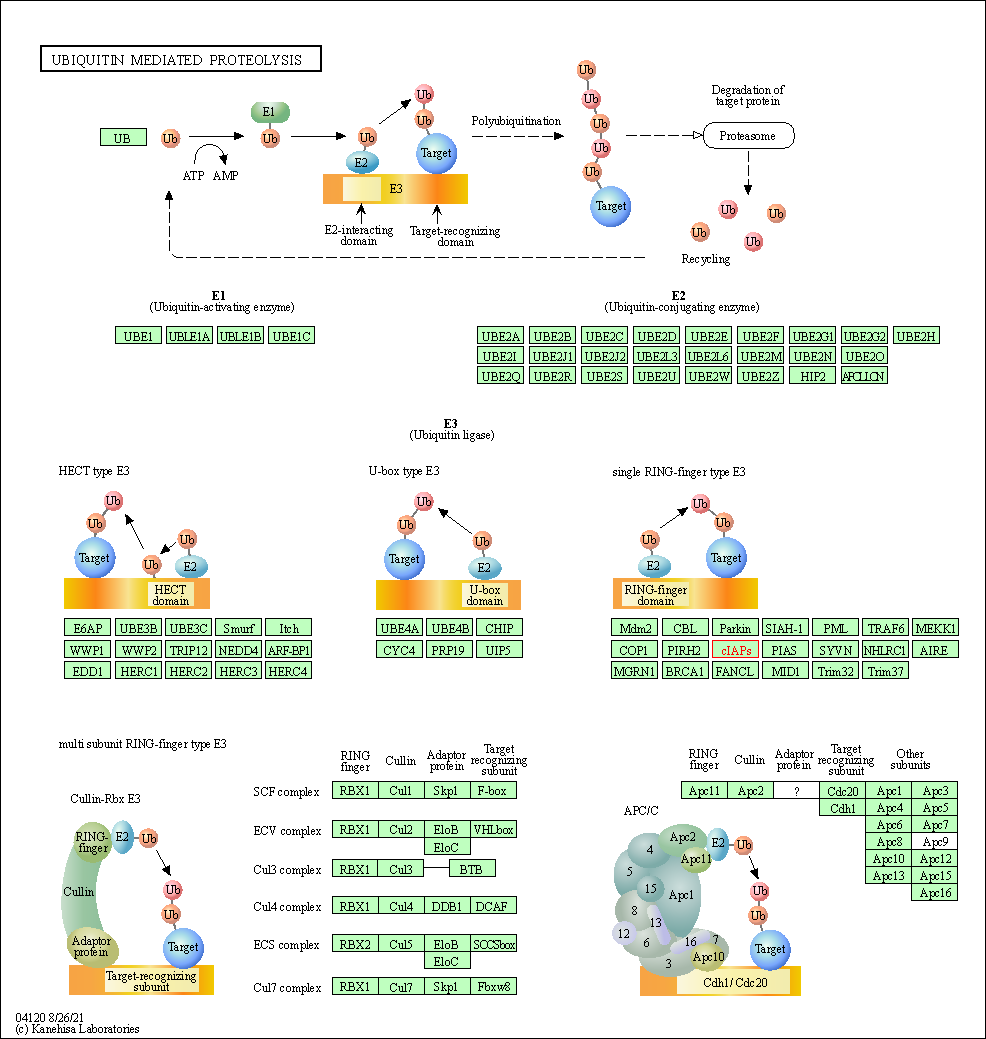
| Ubiquitin mediated proteolysis | hsa04120 | Affiliated Target |

|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
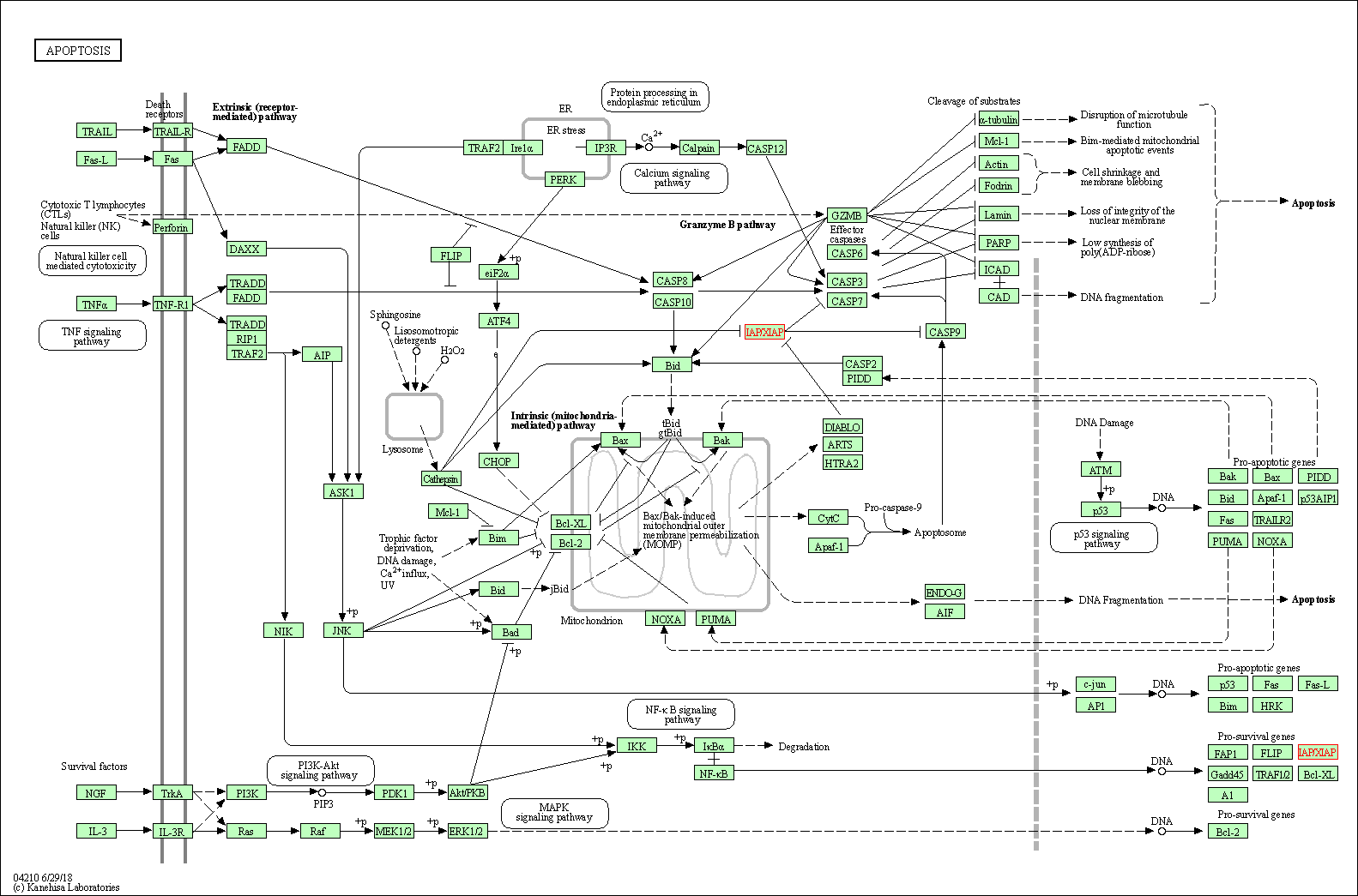
| Apoptosis | hsa04210 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
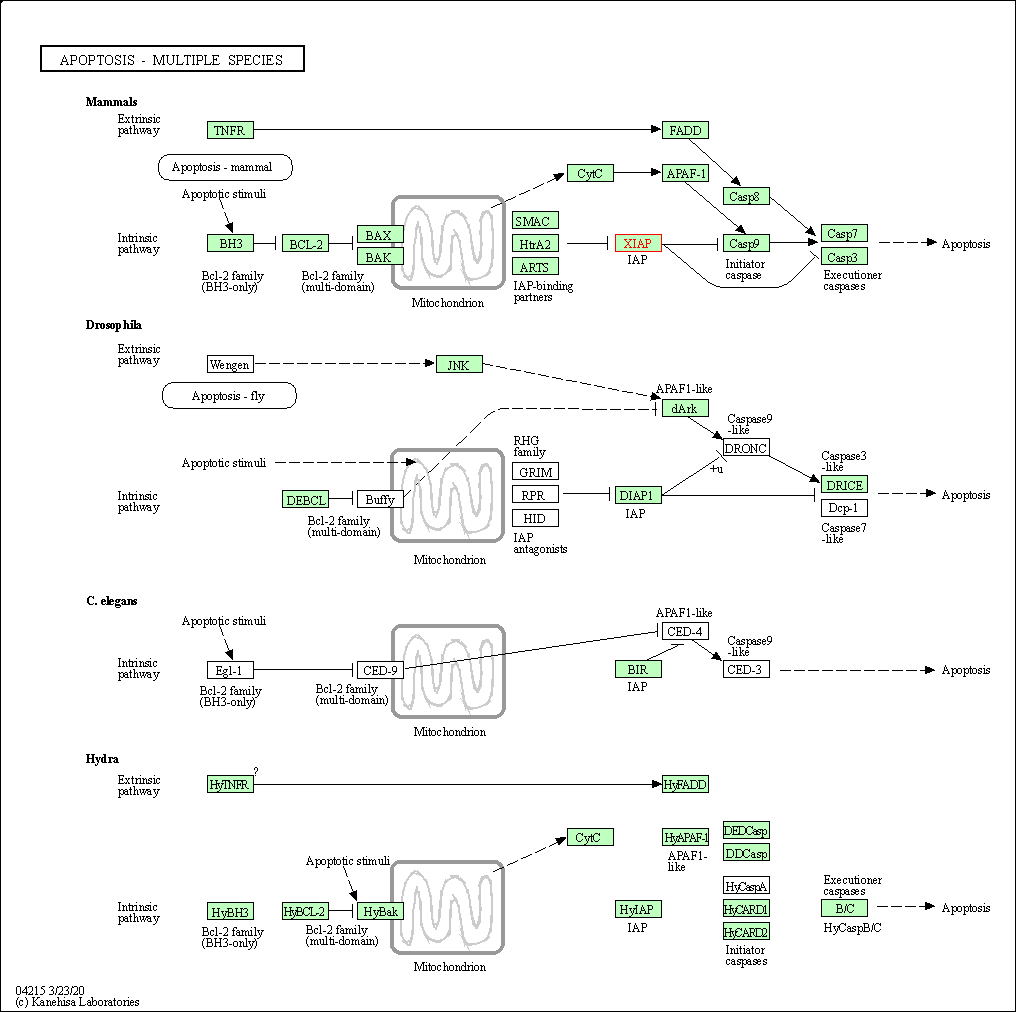
| Apoptosis - multiple species | hsa04215 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
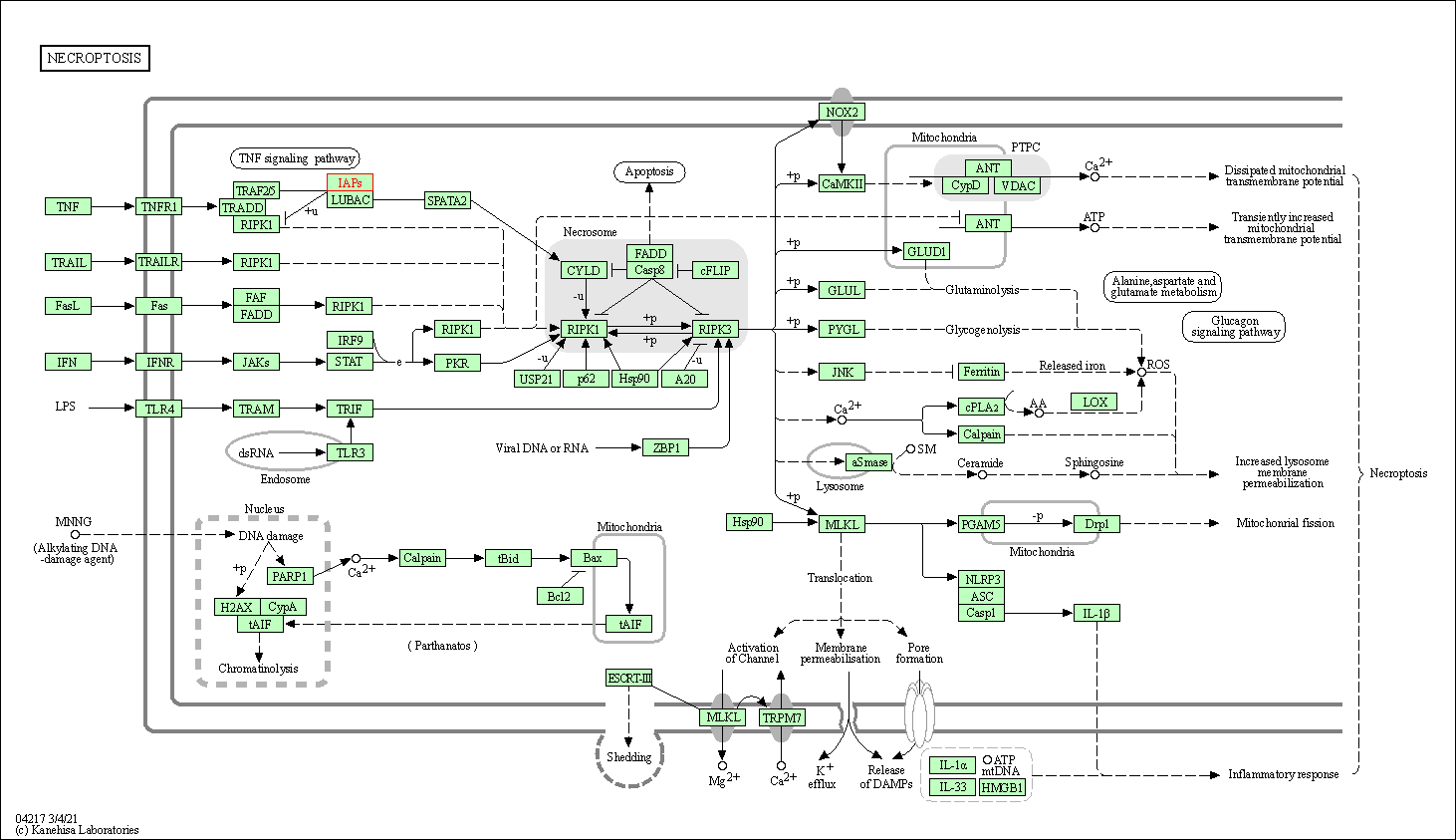
| Necroptosis | hsa04217 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
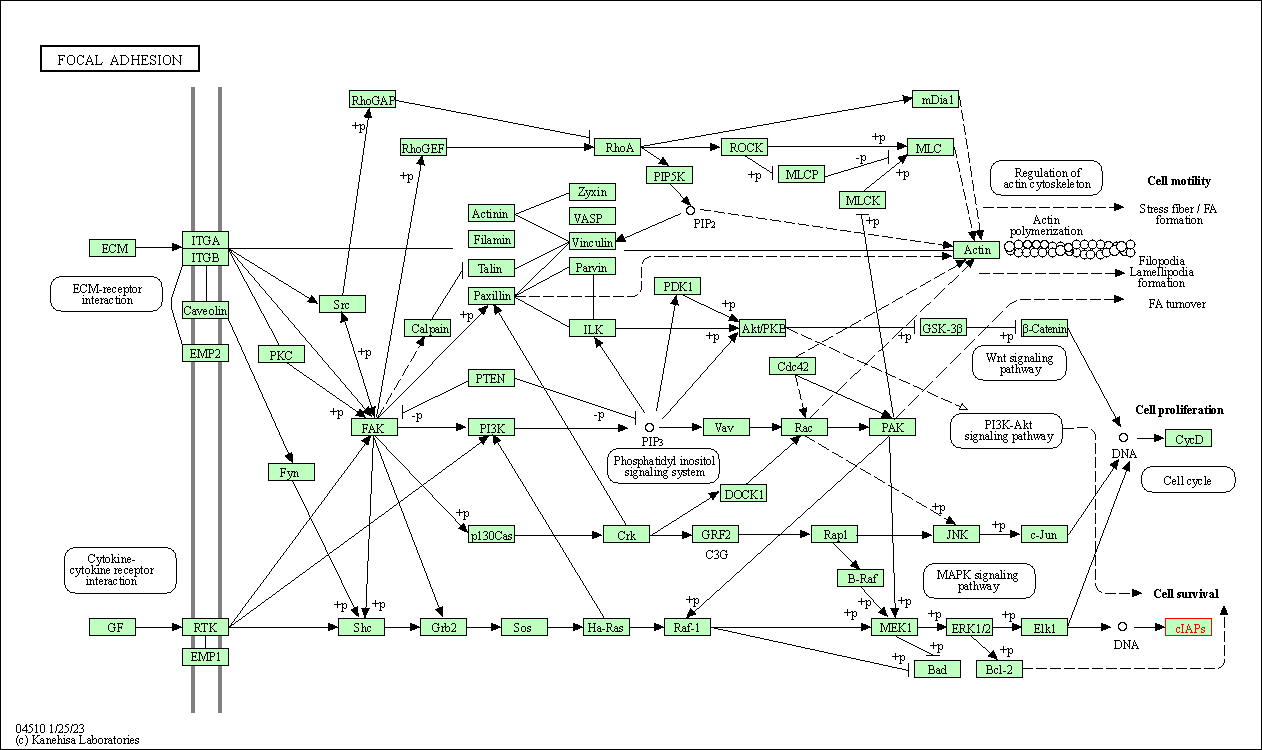
| Focal adhesion | hsa04510 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
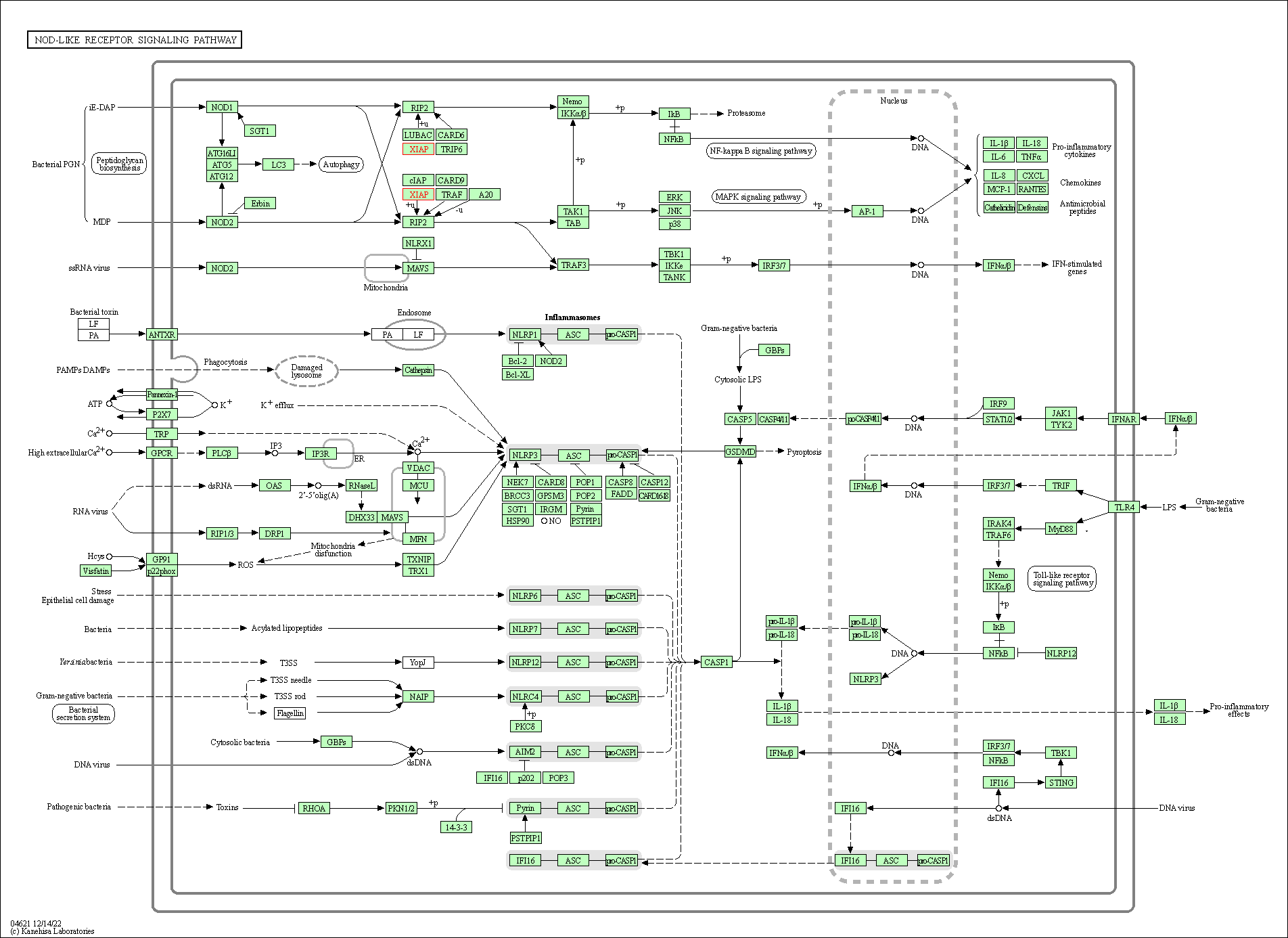
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
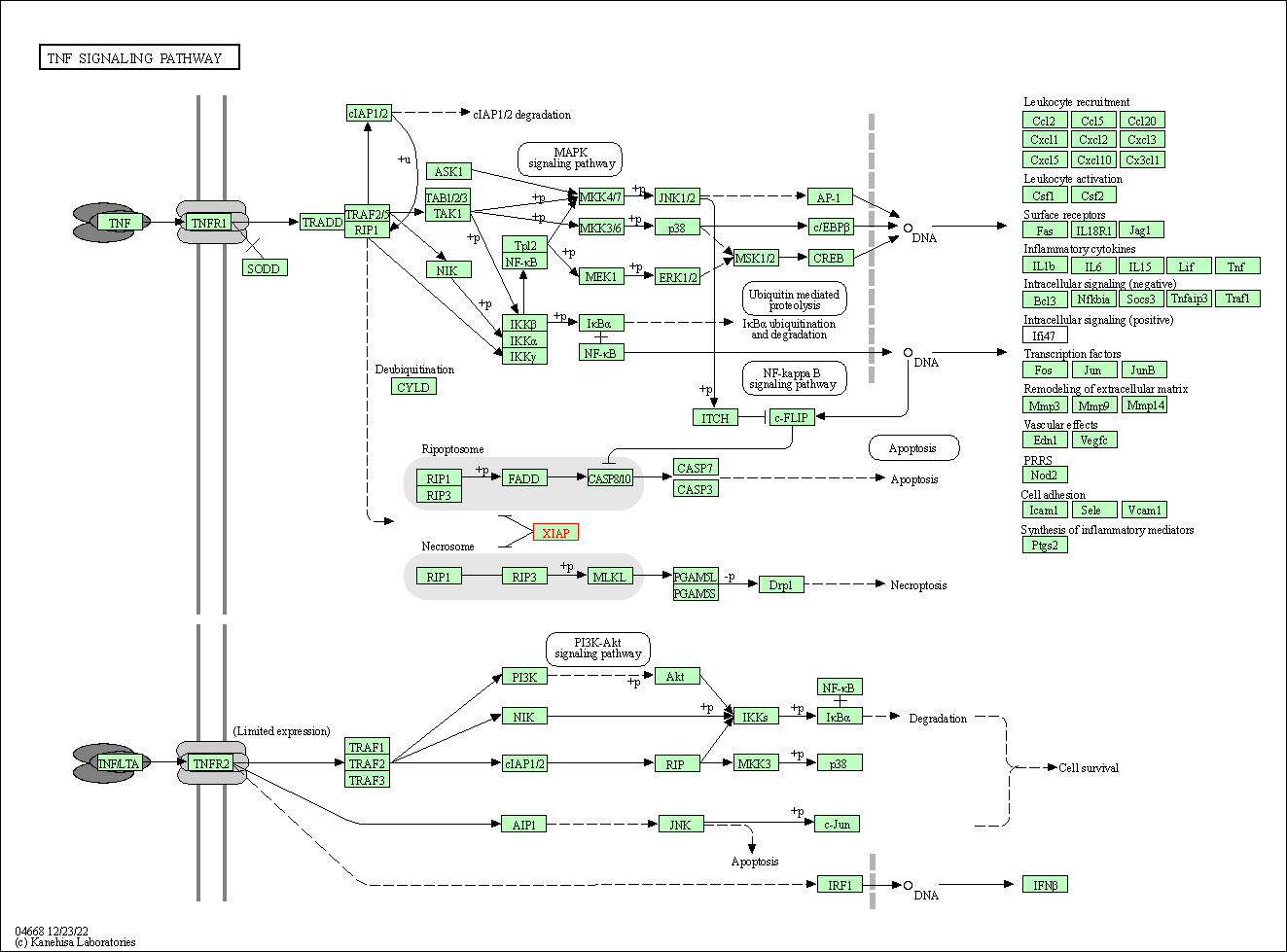
| TNF signaling pathway | hsa04668 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 22 | Degree centrality | 2.36E-03 | Betweenness centrality | 5.24E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.46E-01 | Radiality | 1.43E+01 | Clustering coefficient | 2.25E-01 |
| Neighborhood connectivity | 4.82E+01 | Topological coefficient | 7.47E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 8 KEGG Pathways | + | ||||
| 1 | NF-kappa B signaling pathway | |||||
| 2 | Ubiquitin mediated proteolysis | |||||
| 3 | Apoptosis | |||||
| 4 | Focal adhesion | |||||
| 5 | Toxoplasmosis | |||||
| 6 | HTLV-I infection | |||||
| 7 | Pathways in cancer | |||||
| 8 | Small cell lung cancer | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Apoptosis signaling pathway | |||||
| PID Pathway | [+] 4 PID Pathways | + | ||||
| 1 | p75(NTR)-mediated signaling | |||||
| 2 | BMP receptor signaling | |||||
| 3 | Caspase Cascade in Apoptosis | |||||
| 4 | TGF-beta receptor signaling | |||||
| Reactome | [+] 7 Reactome Pathways | + | ||||
| 1 | SMAC binds to IAPs | |||||
| 2 | SMAC-mediated dissociation of IAP:caspase complexes | |||||
| 3 | Deactivation of the beta-catenin transactivating complex | |||||
| 4 | RIPK1-mediated regulated necrosis | |||||
| 5 | Regulation of TNFR1 signaling | |||||
| 6 | TNFR1-induced NFkappaB signaling pathway | |||||
| 7 | Regulation of necroptotic cell death | |||||
| WikiPathways | [+] 6 WikiPathways | + | ||||
| 1 | Copper homeostasis | |||||
| 2 | Focal Adhesion | |||||
| 3 | Apoptosis | |||||
| 4 | Intrinsic Pathway for Apoptosis | |||||
| 5 | Apoptosis Modulation and Signaling | |||||
| 6 | NOD pathway | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | cIAPs and XIAP regulate myelopoiesis through cytokine production in an RIPK1- and RIPK3-dependent manner. Blood. 2014 Apr 17;123(16):2562-72. | |||||
| REF 2 | ClinicalTrials.gov (NCT04459715) A Study of Debio 1143 (Xevinapant) in Combination With Platinum-Based Chemotherapy and Standard Fractionation Intensity-Modulated Radiotherapy in Participants With Locally Advanced Squamous Cell Carcinoma of the Head and Neck, Suitable for Definitive Chemoradiotherapy. U.S. National Institutes of Health. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7432). | |||||
| REF 4 | ClinicalTrials.gov (NCT02147873) Study of Azacitidine With or Without Birinapant in Subjects With MDS or CMMoL. U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT00382811) OVATURE (OVArian TUmor REsponse) A Phase III Study of Weekly Carboplatin With and Without Phenoxodiol in Patients With Platinum-Resistant, Recurrent Epithelial Ovarian Cancer. U.S. National Institutes of Health. | |||||
| REF 6 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 7 | Discovery of a potent small-molecule antagonist of inhibitor of apoptosis (IAP) proteins and clinical candidate for the treatment of cancer (GDC-0152). J Med Chem. 2012 May 10;55(9):4101-13. | |||||
| REF 8 | ClinicalTrials.gov (NCT00708006) A Study of HGS1029 (AEG40826-2HCl) in Subjects With Advanced Solid Tumors. U.S. National Institutes of Health. | |||||
| REF 9 | Debio 1143, an antagonist of multiple inhibitor-of-apoptosis proteins, activates apoptosis and enhances radiosensitization of non-small cell lung c... Am J Cancer Res. 2014 Nov 19;4(6):943-51. | |||||
| REF 10 | Debio 1143, an antagonist of multiple inhibitor-of-apoptosis proteins, activates apoptosis and enhances radiosensitization of non-small cell lung cancer cells in vitro. Am J Cancer Res. 2014; 4(6): 943-951. | |||||
| REF 11 | Birinapant (TL32711), a bivalent SMAC mimetic, targets TRAF2-associated cIAPs, abrogates TNF-induced NF- B activation, and is active in patient-derived xenograft models. Mol Cancer Ther. 2014 Apr;13(4):867-79. | |||||
| REF 12 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. | |||||
| REF 13 | XIAP Limits Autophagic Degradation of Sox2 and Is A Therapeutic Target in Nasopharyngeal Carcinoma Stem Cells.Theranostics. 2018 Feb 5;8(6):1494-1510. | |||||
| REF 14 | Clinical pipeline report, company report or official report of Aegera Therapeutics Inc. | |||||
| REF 15 | Small molecule inhibitor of apoptosis proteins antagonists: a patent review.Expert Opin Ther Pat. 2015 Jul;25(7):755-74. | |||||
| REF 16 | Design and synthesis of potent inhibitor of apoptosis (IAP) proteins antagonists bearing an octahydropyrrolo[1,2-a]pyrazine scaffold as a novel proline mimetic. J Med Chem. 2013 Feb 14;56(3):1228-46. | |||||
| REF 17 | Structure-based design and molecular profiling of Smac-mimetics selective for cellular IAPs. FEBS J. 2018 Sep;285(17):3286-3298. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

