Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T40954
(Former ID: TTDS00056)
|
|||||
| Target Name |
Xanthine dehydrogenase/oxidase (XDH)
|
|||||
| Synonyms |
Xanthine oxidase; Xanthine dehydrogenase; XDHA
Click to Show/Hide
|
|||||
| Gene Name |
XDH
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Inborn purine/pyrimidine/nucleotide metabolism error [ICD-11: 5C55] | |||||
| 2 | Mineral deficiency [ICD-11: 5B5K] | |||||
| Function |
Catalyzes the oxidation of hypoxanthine to xanthine. Catalyzes the oxidation of xanthine to uric acid. Contributes to the generation of reactive oxygen species. Has also low oxidase activity towards aldehydes (in vitro). Key enzyme in purine degradation.
Click to Show/Hide
|
|||||
| BioChemical Class |
CH/CH(2) oxidoreductase
|
|||||
| UniProt ID | ||||||
| Sequence |
MTADKLVFFVNGRKVVEKNADPETTLLAYLRRKLGLSGTKLGCGEGGCGACTVMLSKYDR
LQNKIVHFSANACLAPICSLHHVAVTTVEGIGSTKTRLHPVQERIAKSHGSQCGFCTPGI VMSMYTLLRNQPEPTMEEIENAFQGNLCRCTGYRPILQGFRTFARDGGCCGGDGNNPNCC MNQKKDHSVSLSPSLFKPEEFTPLDPTQEPIFPPELLRLKDTPRKQLRFEGERVTWIQAS TLKELLDLKAQHPDAKLVVGNTEIGIEMKFKNMLFPMIVCPAWIPELNSVEHGPDGISFG AACPLSIVEKTLVDAVAKLPAQKTEVFRGVLEQLRWFAGKQVKSVASVGGNIITASPISD LNPVFMASGAKLTLVSRGTRRTVQMDHTFFPGYRKTLLSPEEILLSIEIPYSREGEYFSA FKQASRREDDIAKVTSGMRVLFKPGTTEVQELALCYGGMANRTISALKTTQRQLSKLWKE ELLQDVCAGLAEELHLPPDAPGGMVDFRCTLTLSFFFKFYLTVLQKLGQENLEDKCGKLD PTFASATLLFQKDPPADVQLFQEVPKGQSEEDMVGRPLPHLAADMQASGEAVYCDDIPRY ENELSLRLVTSTRAHAKIKSIDTSEAKKVPGFVCFISADDVPGSNITGICNDETVFAKDK VTCVGHIIGAVVADTPEHTQRAAQGVKITYEELPAIITIEDAIKNNSFYGPELKIEKGDL KKGFSEADNVVSGEIYIGGQEHFYLETHCTIAVPKGEAGEMELFVSTQNTMKTQSFVAKM LGVPANRIVVRVKRMGGGFGGKETRSTVVSTAVALAAYKTGRPVRCMLDRDEDMLITGGR HPFLARYKVGFMKTGTVVALEVDHFSNVGNTQDLSQSIMERALFHMDNCYKIPNIRGTGR LCKTNLPSNTAFRGFGGPQGMLIAECWMSEVAVTCGMPAEEVRRKNLYKEGDLTHFNQKL EGFTLPRCWEECLASSQYHARKSEVDKFNKENCWKKRGLCIIPTKFGISFTVPFLNQAGA LLHVYTDGSVLLTHGGTEMGQGLHTKMVQVASRALKIPTSKIYISETSTNTVPNTSPTAA SVSADLNGQAVYAACQTILKRLEPYKKKNPSGSWEDWVTAAYMDTVSLSATGFYRTPNLG YSFETNSGNPFHYFSYGVACSEVEIDCLTGDHKNLRTDIVMDVGSSLNPAIDIGQVEGAF VQGLGLFTLEELHYSPEGSLHTRGPSTYKIPAFGSIPIEFRVSLLRDCPNKKAIYASKAV GEPPLFLAASIFFAIKDAIRAARAQHTGNNVKELFRLDSPATPEKIRNACVDKFTTLCVT GVPENCKPWSVRV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T87DEX | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 3 Approved Drugs | + | ||||
| 1 | Allopurinol | Drug Info | Approved | Hyperuricaemia | [2], [3] | |
| 2 | Febuxostat | Drug Info | Approved | Hyperuricaemia | [4], [5] | |
| 3 | Fosdenopterin | Drug Info | Approved | Molybdenum cofactor deficiency | [6] | |
| Clinical Trial Drug(s) | [+] 6 Clinical Trial Drugs | + | ||||
| 1 | Curcumin | Drug Info | Phase 3 | Solid tumour/cancer | [7], [8] | |
| 2 | Oxypurinol | Drug Info | Phase 2/3 | Heart failure | [9] | |
| 3 | BAICALEIN | Drug Info | Phase 2 | Influenza virus infection | [10] | |
| 4 | TMX-049 | Drug Info | Phase 2 | Diabetic nephropathy | [11] | |
| 5 | Topiroxostat | Drug Info | Phase 2 | Gout | [12] | |
| 6 | LC-350189 | Drug Info | Phase 1 | Gout | [13] | |
| Discontinued Drug(s) | [+] 2 Discontinued Drugs | + | ||||
| 1 | BOF-4272 | Drug Info | Discontinued in Phase 2 | Gout | [14] | |
| 2 | Y-700 | Drug Info | Discontinued in Phase 2 | Gout | [15] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 66 Inhibitor drugs | + | ||||
| 1 | Allopurinol | Drug Info | [1], [16], [17] | |||
| 2 | Febuxostat | Drug Info | [18], [5] | |||
| 3 | Curcumin | Drug Info | [19] | |||
| 4 | Oxypurinol | Drug Info | [20] | |||
| 5 | BAICALEIN | Drug Info | [21] | |||
| 6 | TMX-049 | Drug Info | [22] | |||
| 7 | Topiroxostat | Drug Info | [23] | |||
| 8 | LC-350189 | Drug Info | [24] | |||
| 9 | Azaindole derivative 3 | Drug Info | [25] | |||
| 10 | Azaindole derivative 4 | Drug Info | [25] | |||
| 11 | Azaindole derivative 5 | Drug Info | [25] | |||
| 12 | Azaindole derivative 6 | Drug Info | [25] | |||
| 13 | Azaindole derivative 7 | Drug Info | [25] | |||
| 14 | Azole benzene derivative 1 | Drug Info | [25] | |||
| 15 | Azole benzene derivative 2 | Drug Info | [25] | |||
| 16 | Azole benzene derivative 3 | Drug Info | [25] | |||
| 17 | Azole benzene derivative 4 | Drug Info | [25] | |||
| 18 | Fused heterocyclic compound 1 | Drug Info | [25] | |||
| 19 | Fused heterocyclic compound 10 | Drug Info | [25] | |||
| 20 | Fused heterocyclic compound 11 | Drug Info | [25] | |||
| 21 | Fused heterocyclic compound 2 | Drug Info | [25] | |||
| 22 | Fused heterocyclic compound 3 | Drug Info | [25] | |||
| 23 | Fused heterocyclic compound 4 | Drug Info | [25] | |||
| 24 | Fused heterocyclic compound 5 | Drug Info | [25] | |||
| 25 | Fused heterocyclic compound 6 | Drug Info | [25] | |||
| 26 | Fused heterocyclic compound 7 | Drug Info | [25] | |||
| 27 | Fused heterocyclic compound 8 | Drug Info | [25] | |||
| 28 | Fused heterocyclic compound 9 | Drug Info | [25] | |||
| 29 | PMID27841045-Compound-129 | Drug Info | [25] | |||
| 30 | PMID27841045-Compound-130 | Drug Info | [25] | |||
| 31 | PMID27841045-Compound-131 | Drug Info | [25] | |||
| 32 | PMID27841045-Compound-132 | Drug Info | [25] | |||
| 33 | PMID27841045-Compound-133 | Drug Info | [25] | |||
| 34 | PMID27841045-Compound-134 | Drug Info | [25] | |||
| 35 | PMID27841045-Compound-135 | Drug Info | [25] | |||
| 36 | PMID27841045-Compound-136 | Drug Info | [25] | |||
| 37 | PMID27841045-Compound-137 | Drug Info | [25] | |||
| 38 | PMID27841045-Compound-143 | Drug Info | [25] | |||
| 39 | PMID27841045-Compound-144 | Drug Info | [25] | |||
| 40 | PMID27841045-Compound-145 | Drug Info | [25] | |||
| 41 | PMID27841045-Compound-155 | Drug Info | [25] | |||
| 42 | PMID27841045-Compound-156 | Drug Info | [25] | |||
| 43 | PMID27841045-Compound-157 | Drug Info | [25] | |||
| 44 | BOF-4272 | Drug Info | [26], [27] | |||
| 45 | Y-700 | Drug Info | [28] | |||
| 46 | 1,2,3,4,6-penta-O-galloyl-beta-D-glucose | Drug Info | [29] | |||
| 47 | 1-(3-Cyano-phenyl)-1H-pyrazole-4-carboxylic acid | Drug Info | [30] | |||
| 48 | 2-chloro-6(methylamino)purine | Drug Info | [31] | |||
| 49 | 3,4'-(1H-1,2,4-triazole-3,5-diyl)dipyridine | Drug Info | [32] | |||
| 50 | 3,5-bis(2-methylpyridin-4-yl)-1H-1,2,4-triazole | Drug Info | [32] | |||
| 51 | 3,5-di(pyridin-4-yl)-1H-1,2,4-triazole | Drug Info | [32] | |||
| 52 | 3-O-METHYLQUERCETIN | Drug Info | [21] | |||
| 53 | ACACETIN | Drug Info | [21] | |||
| 54 | AMENTOFLAVONE | Drug Info | [21] | |||
| 55 | Aom-0763 | Drug Info | [33] | |||
| 56 | CHRYSOERIOL | Drug Info | [21] | |||
| 57 | DIHYDROKAEMPFEROL | Drug Info | [21] | |||
| 58 | Dioxothiomolybdenum(VI) ion | Drug Info | [34] | |||
| 59 | Flavin-Adenine Dinucleotide | Drug Info | [35] | |||
| 60 | FUKUGETIN | Drug Info | [21] | |||
| 61 | LIQUIRTIGENIN | Drug Info | [21] | |||
| 62 | NC-2500 | Drug Info | [33] | |||
| 63 | PERSICARIN | Drug Info | [21] | |||
| 64 | ROBINETIN | Drug Info | [21] | |||
| 65 | SCUTELLAREIN | Drug Info | [21] | |||
| 66 | Wogonin | Drug Info | [21] | |||
| Cofactor | [+] 1 Cofactor drugs | + | ||||
| 1 | Fosdenopterin | Drug Info | [6] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Salicyclic acid | Ligand Info | |||||
| Structure Description | Crystal Structure of Human Xanthine Oxidoreductase mutant, Glu803Val | PDB:2E1Q | ||||
| Method | X-ray diffraction | Resolution | 2.60 Å | Mutation | Yes | [36] |
| PDB Sequence |
ADKLVFFVNG
12 RKVVEKNADP22 ETTLLAYLRR32 KLGLSGTKLG42 CGEGGCGACT52 VMLSKYDRLQ 62 NKIVHFSANA72 CLAPICSLHH82 VAVTTVEGIG92 STKTRLHPVQ102 ERIAKSHGSQ 112 CGFCTPGIVM122 SMYTLLRNQP132 EPTMEEIENA142 FQGNLCRCTG152 YRPILQGFRT 162 FARDGSPSLF196 KPEEFTPLDP206 TQEPIFPPEL216 LRLKDTPRKQ226 LRFEGERVTW 236 IQASTLKELL246 DLKAQHPDAK256 LVVGNTEIGI266 EMKFKNMLFP276 MIVCPAWIPE 286 LNSVEHGPDG296 ISFGAACPLS306 IVEKTLVDAV316 AKLPAQKTEV326 FRGVLEQLRW 336 FAGKQVKSVA346 SVGGNIITAS356 PISDLNPVFM366 ASGAKLTLVS376 RGTRRTVQMD 386 HTFFPGYRKT396 LLSPEEILLS406 IEIPYSREGE416 YFSAFKQASR426 REDDIAKVTS 436 GMRVLFKPGT446 TEVQELALCY456 GGMANRTISA466 LKTTQRQLSK476 LWKEELLQDV 486 CAGLAEELHL496 PPDAPGGMVD506 FRCTLTLSFF516 FKFYLTVLQK526 LGQENLEDKC 536 GKLDPTFASA546 TLLFQKDPPA556 DVQLFQEVPK566 GQSEEDMVGR576 PLPHLAADMQ 586 ASGEAVYCDD596 IPRYENELSL606 RLVTSTRAHA616 KIKSIDTSEA626 KKVPGFVCFI 636 SADDVPGSNI646 TGICNDETVF656 AKDKVTCVGH666 IIGAVVADTP676 EHTQRAAQGV 686 KITYEELPAI696 ITIEDAIKNN706 SFYGPELKIE716 KGDLKKGFSE726 ADNVVSGEIY 736 IGGQEHFYLE746 THCTIAVPKG756 EAGEMELFVS766 TQNTMKTQSF776 VAKMLGVPAN 786 RIVVRVKRMG796 GGFGGKVTRS806 TVVSTAVALA816 AYKTGRPVRC826 MLDRDEDMLI 836 TGGRHPFLAR846 YKVGFMKTGT856 VVALEVDHFS866 NVGNTQDLSQ876 SIMERALFHM 886 DNCYKIPNIR896 GTGRLCKTNL906 PSNTAFRGFG916 GPQGMLIAEC926 WMSEVAVTCG 936 MPAEEVRRKN946 LYKEGDLTHF956 NQKLEGFTLP966 RCWEECLASS976 QYHARKSEVD 986 KFNKENCWKK996 RGLCIIPTKF1006 GISFTVPFLN1016 QAGALLHVYT1026 DGSVLLTHGG 1036 TEMGQGLHTK1046 MVQVASRALK1056 IPTSKIYISE1066 TSTNTVPNTS1076 PTAASVSADL 1086 NGQAVYAACQ1096 TILKRLEPYK1106 KKNPSGSWED1116 WVTAAYMDTV1126 SLSATGFYRT 1136 PNLGYSFETN1146 SGNPFHYFSY1156 GVACSEVEID1166 CLTGDHKNLR1176 TDIVMDVGSS 1186 LNPAIDIGQV1196 EGAFVQGLGL1206 FTLEELHYSP1216 EGSLHTRGPS1226 TYKIPAFGSI 1236 PIEFRVSLLR1246 DCPNKKAIYA1256 SKAVGEPPLF1266 LAASIFFAIK1276 DAIRAARAQH 1286 TGNNVKELFR1296 LDSPATPEKI1306 RNACVDKFTT1316 LCVTGVPENC1326 KPWSVRV |
|||||
|
|
||||||
| Ligand Name: Flavin-Adenine Dinucleotide | Ligand Info | |||||
| Structure Description | Crystal Structure of Human Xanthine Oxidoreductase mutant, Glu803Val | PDB:2E1Q | ||||
| Method | X-ray diffraction | Resolution | 2.60 Å | Mutation | Yes | [36] |
| PDB Sequence |
ADKLVFFVNG
12 RKVVEKNADP22 ETTLLAYLRR32 KLGLSGTKLG42 CGEGGCGACT52 VMLSKYDRLQ 62 NKIVHFSANA72 CLAPICSLHH82 VAVTTVEGIG92 STKTRLHPVQ102 ERIAKSHGSQ 112 CGFCTPGIVM122 SMYTLLRNQP132 EPTMEEIENA142 FQGNLCRCTG152 YRPILQGFRT 162 FARDGSPSLF196 KPEEFTPLDP206 TQEPIFPPEL216 LRLKDTPRKQ226 LRFEGERVTW 236 IQASTLKELL246 DLKAQHPDAK256 LVVGNTEIGI266 EMKFKNMLFP276 MIVCPAWIPE 286 LNSVEHGPDG296 ISFGAACPLS306 IVEKTLVDAV316 AKLPAQKTEV326 FRGVLEQLRW 336 FAGKQVKSVA346 SVGGNIITAS356 PISDLNPVFM366 ASGAKLTLVS376 RGTRRTVQMD 386 HTFFPGYRKT396 LLSPEEILLS406 IEIPYSREGE416 YFSAFKQASR426 REDDIAKVTS 436 GMRVLFKPGT446 TEVQELALCY456 GGMANRTISA466 LKTTQRQLSK476 LWKEELLQDV 486 CAGLAEELHL496 PPDAPGGMVD506 FRCTLTLSFF516 FKFYLTVLQK526 LGQENLEDKC 536 GKLDPTFASA546 TLLFQKDPPA556 DVQLFQEVPK566 GQSEEDMVGR576 PLPHLAADMQ 586 ASGEAVYCDD596 IPRYENELSL606 RLVTSTRAHA616 KIKSIDTSEA626 KKVPGFVCFI 636 SADDVPGSNI646 TGICNDETVF656 AKDKVTCVGH666 IIGAVVADTP676 EHTQRAAQGV 686 KITYEELPAI696 ITIEDAIKNN706 SFYGPELKIE716 KGDLKKGFSE726 ADNVVSGEIY 736 IGGQEHFYLE746 THCTIAVPKG756 EAGEMELFVS766 TQNTMKTQSF776 VAKMLGVPAN 786 RIVVRVKRMG796 GGFGGKVTRS806 TVVSTAVALA816 AYKTGRPVRC826 MLDRDEDMLI 836 TGGRHPFLAR846 YKVGFMKTGT856 VVALEVDHFS866 NVGNTQDLSQ876 SIMERALFHM 886 DNCYKIPNIR896 GTGRLCKTNL906 PSNTAFRGFG916 GPQGMLIAEC926 WMSEVAVTCG 936 MPAEEVRRKN946 LYKEGDLTHF956 NQKLEGFTLP966 RCWEECLASS976 QYHARKSEVD 986 KFNKENCWKK996 RGLCIIPTKF1006 GISFTVPFLN1016 QAGALLHVYT1026 DGSVLLTHGG 1036 TEMGQGLHTK1046 MVQVASRALK1056 IPTSKIYISE1066 TSTNTVPNTS1076 PTAASVSADL 1086 NGQAVYAACQ1096 TILKRLEPYK1106 KKNPSGSWED1116 WVTAAYMDTV1126 SLSATGFYRT 1136 PNLGYSFETN1146 SGNPFHYFSY1156 GVACSEVEID1166 CLTGDHKNLR1176 TDIVMDVGSS 1186 LNPAIDIGQV1196 EGAFVQGLGL1206 FTLEELHYSP1216 EGSLHTRGPS1226 TYKIPAFGSI 1236 PIEFRVSLLR1246 DCPNKKAIYA1256 SKAVGEPPLF1266 LAASIFFAIK1276 DAIRAARAQH 1286 TGNNVKELFR1296 LDSPATPEKI1306 RNACVDKFTT1316 LCVTGVPENC1326 KPWSVRV |
|||||
|
|
GLU45
3.478
GLY46
2.959
GLY47
3.816
LEU74
3.733
LYS256
3.220
LEU257
2.805
VAL258
3.418
VAL259
2.673
GLY260
2.972
ASN261
2.844
THR262
2.557
GLU263
2.987
ILE264
2.940
GLY265
4.765
ILE266
4.232
GLU267
4.949
LEU287
4.456
ALA301
3.892
LEU305
4.148
TRP336
3.933
PHE337
3.199
ALA338
3.403
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Purine metabolism | hsa00230 | Affiliated Target |
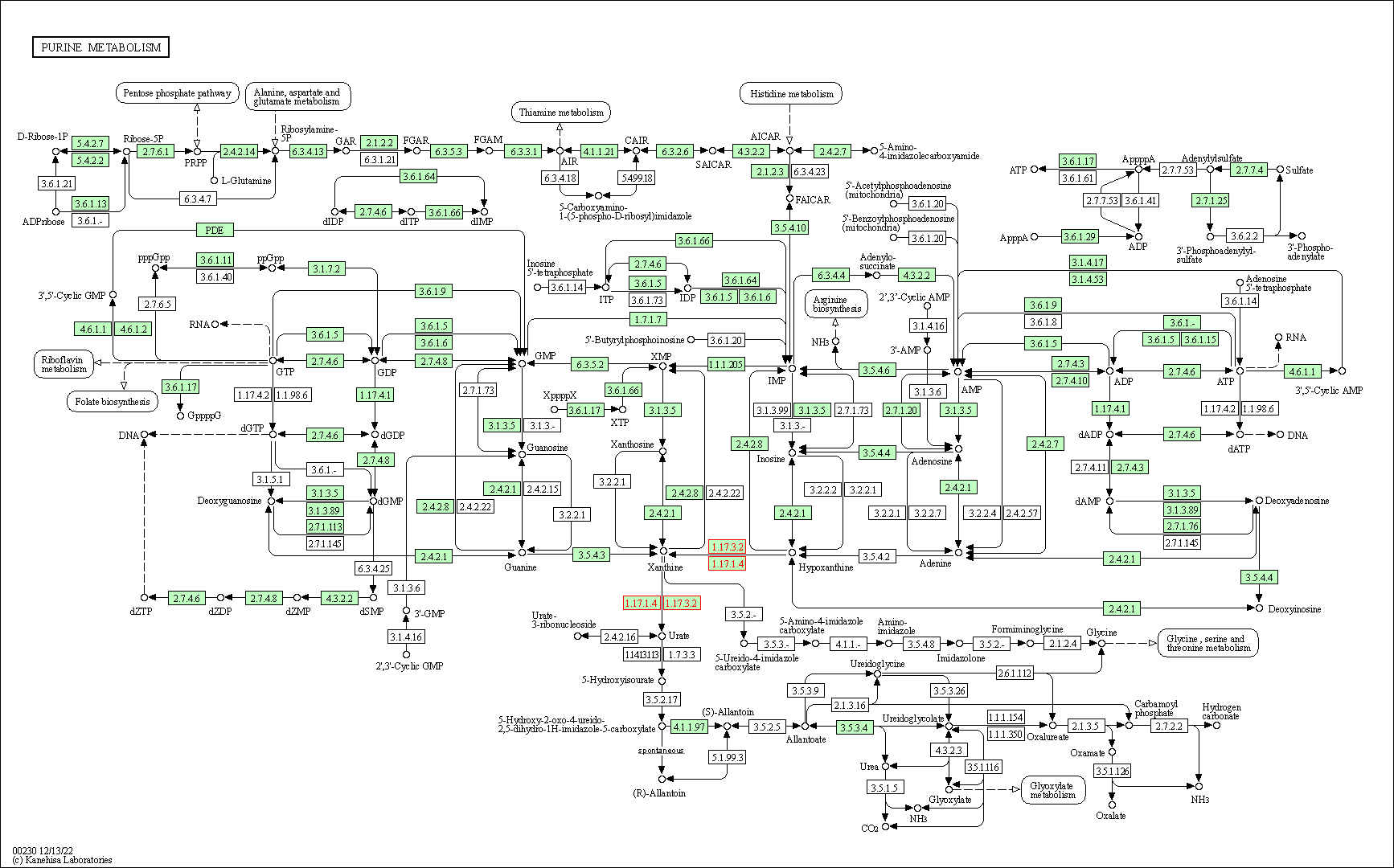
|
| Class: Metabolism => Nucleotide metabolism | Pathway Hierarchy | ||
| Caffeine metabolism | hsa00232 | Affiliated Target |
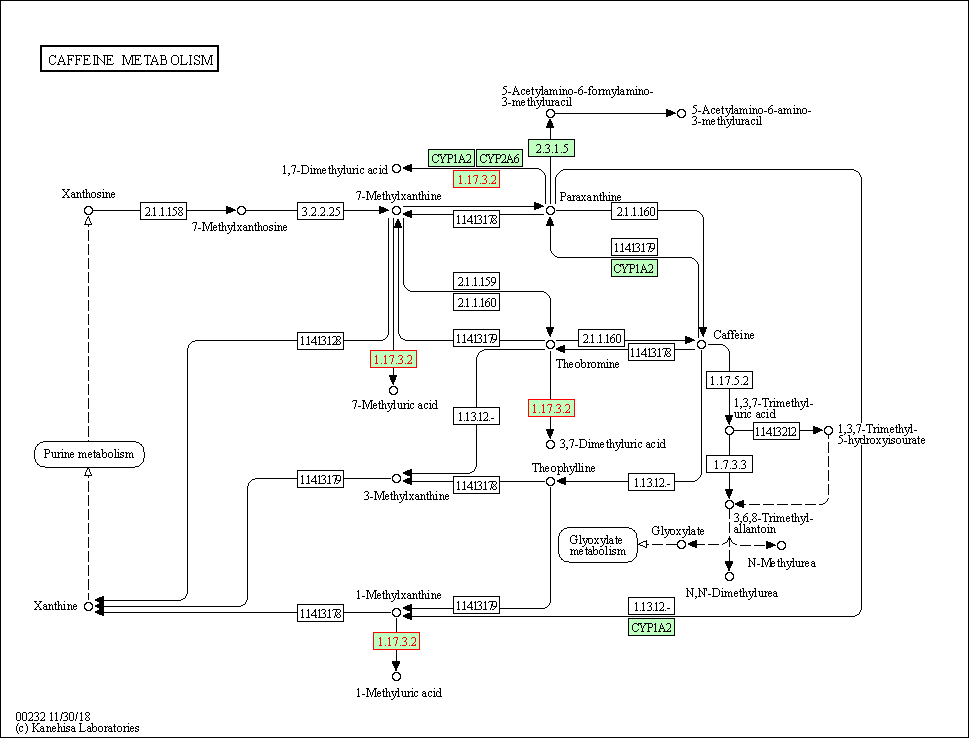
|
| Class: Metabolism => Biosynthesis of other secondary metabolites | Pathway Hierarchy | ||
| Drug metabolism - other enzymes | hsa00983 | Affiliated Target |
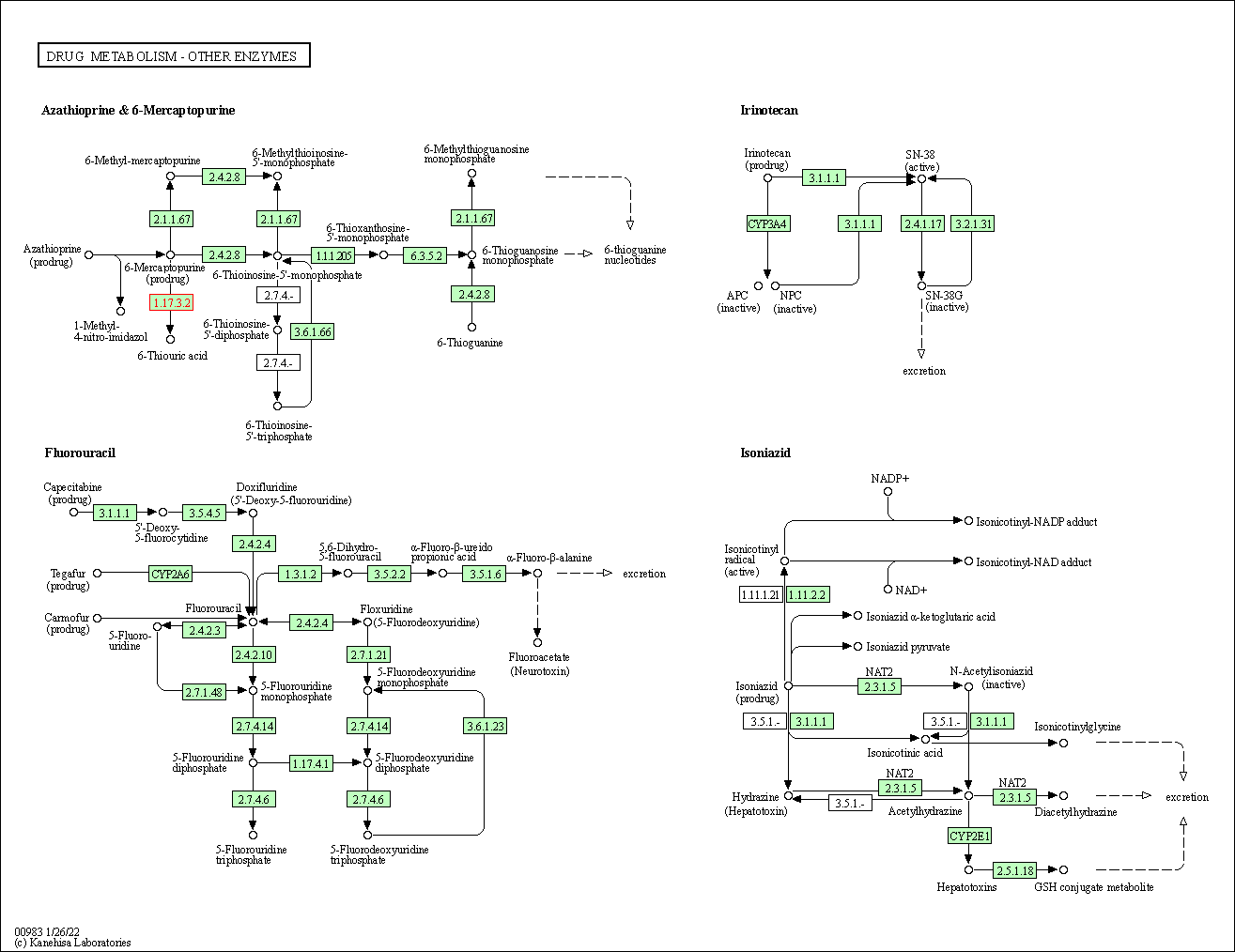
|
| Class: Metabolism => Xenobiotics biodegradation and metabolism | Pathway Hierarchy | ||
| Peroxisome | hsa04146 | Affiliated Target |
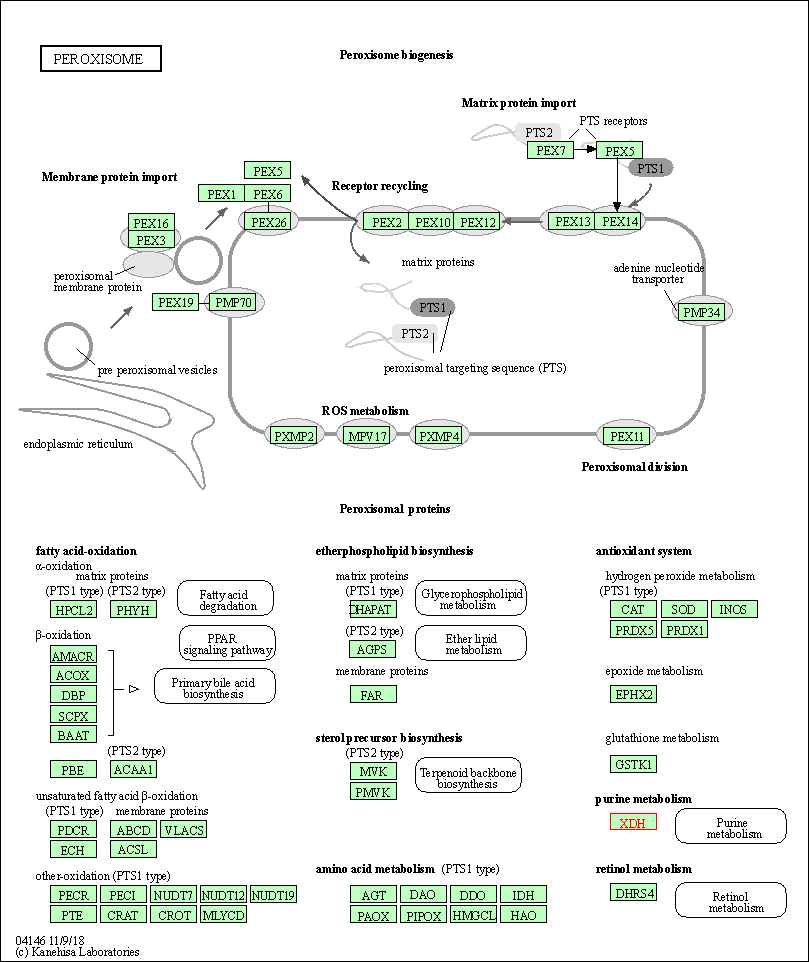
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Degree | 2 | Degree centrality | 2.15E-04 | Betweenness centrality | 2.02E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.38E-01 | Radiality | 1.15E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 2.50E+00 | Topological coefficient | 5.00E-01 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 5 BioCyc Pathways | + | ||||
| 1 | Purine nucleotides degradation | |||||
| 2 | Urate biosynthesis/inosine 5'-phosphate degradation | |||||
| 3 | Guanosine nucleotides degradation | |||||
| 4 | Adenosine nucleotides degradation | |||||
| 5 | Retinoate biosynthesis II | |||||
| KEGG Pathway | [+] 5 KEGG Pathways | + | ||||
| 1 | Purine metabolism | |||||
| 2 | Caffeine metabolism | |||||
| 3 | Drug metabolism - other enzymes | |||||
| 4 | Metabolic pathways | |||||
| 5 | Peroxisome | |||||
| Panther Pathway | [+] 2 Panther Pathways | + | ||||
| 1 | Adenine and hypoxanthine salvage pathway | |||||
| 2 | Purine metabolism | |||||
| Pathwhiz Pathway | [+] 2 Pathwhiz Pathways | + | ||||
| 1 | Caffeine Metabolism | |||||
| 2 | Purine Metabolism | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Purine catabolism | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Oxidative Stress | |||||
| 2 | Effects of Nitric Oxide | |||||
| 3 | Metabolism of nucleotides | |||||
| 4 | Selenium Micronutrient Network | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| Target QSAR Model | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Allopurinol: xanthine oxidase inhibitor. Tex Med. 1966 Jan;62(1):100-1. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6795). | |||||
| REF 3 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 018659. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6817). | |||||
| REF 5 | Hughes B: 2009 FDA drug approvals. Nat Rev Drug Discov. 2010 Feb;9(2):89-92. | |||||
| REF 6 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health Human Services. 2021 | |||||
| REF 7 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7000). | |||||
| REF 8 | Nanocurcumin: a promising therapeutic advancement over native curcumin. Crit Rev Ther Drug Carrier Syst. 2013;30(4):331-68. | |||||
| REF 9 | ClinicalTrials.gov (NCT00063687) Oxypurinol Compared With Placebo for Class III-IV NYHA Congestive Heart Failure. U.S. National Institutes of Health. | |||||
| REF 10 | ClinicalTrials.gov (NCT03830684) A Randomized, Double-blind, Placebo-controlled, Multicenter and Phase IIa Clinical Trial for the Effectiveness and Safety of Baicalein Tablets in the Treatment of Improve Other Aspects of Healthy Adult With Influenza Fever. U.S. National Institutes of Health. | |||||
| REF 11 | ClinicalTrials.gov (NCT03449199) Phase 2 Study of TMX-049 in Subjects With Type 2 Diabetes and Albuminuria. U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT02327754) Effect of Topiroxostat on Urinary Albumin Excretion Early Stage Diabetic Nephropathy and Hyperuricemia With or Without Gout. U.S. National Institutes of Health. | |||||
| REF 13 | ClinicalTrials.gov (NCT01361646) Safety, Tolerability and Pharmacokinetic/Pharmacodynamic Characteristics of LC350189. U.S. National Institutes of Health. | |||||
| REF 14 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800002649) | |||||
| REF 15 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800010582) | |||||
| REF 16 | Purification, crystallization and preliminary X-ray diffraction studies of xanthine dehydrogenase and xanthine oxidase isolated from bovine milk. Acta Crystallogr D Biol Crystallogr. 2000 Dec;56(Pt 12):1656-8. | |||||
| REF 17 | Influence of postischemic administration of oxyradical antagonists on ischemic injury to rabbit skeletal muscle. Microsurgery. 1996;17(9):517-23. | |||||
| REF 18 | Clinical pipeline report, company report or official report of Takeda (2009). | |||||
| REF 19 | Inhibition studies of bovine xanthine oxidase by luteolin, silibinin, quercetin, and curcumin. J Nat Prod. 2009 Apr;72(4):725-31. | |||||
| REF 20 | Oxypurinol as an inhibitor of xanthine oxidase-catalyzed production of superoxide radical. Biochem Pharmacol. 1988 Jan 15;37(2):349-52. | |||||
| REF 21 | Inhibition of cow's milk xanthine oxidase by flavonoids. J Nat Prod. 1988 Mar-Apr;51(2):345-8. | |||||
| REF 22 | Clinical pipeline report, company report or official report of Teijin Pharma. | |||||
| REF 23 | QT/QTc study conducted in Japanese adult healthy subjects: a novel xanthine oxidase inhibitor topiroxostat was not associated with QT prolongation. J Clin Pharmacol. 2014 Apr;54(4):446-52. | |||||
| REF 24 | Pharmacokinetics, pharmacodynamics, and tolerability of LC350189, a novel xanthine oxidase inhibitor, in healthy subjects. Drug Des Devel Ther. 2015 Aug 31;9:5033-49. | |||||
| REF 25 | An updated patent review: xanthine oxidase inhibitors for the treatment of hyperuricemia and gout (2011-2015).Expert Opin Ther Pat. 2017 Mar;27(3):311-345. | |||||
| REF 26 | Enantioselective uptake of BOF-4272, a xanthine oxidase inhibitor with a chiral sulfoxide, by isolated rat hepatocytes. Yakugaku Zasshi. 2001 Dec;121(12):989-94. | |||||
| REF 27 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 28 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 29 | Pentagalloylglucose, a xanthine oxidase inhibitor from a Paraguayan crude drug, "Molle-i" (Schinus terebinthifolius). J Nat Prod. 1989 Jan-Feb;52(1):210-1. | |||||
| REF 30 | Synthesis and structure-activity relationships of 1-phenylpyrazoles as xanthine oxidase inhibitors. Bioorg Med Chem Lett. 2001 Apr 9;11(7):879-82. | |||||
| REF 31 | The screening and characterization of 6-aminopurine-based xanthine oxidase inhibitors. Bioorg Med Chem. 2007 May 15;15(10):3450-6. | |||||
| REF 32 | Discovery of 3-(2-cyano-4-pyridyl)-5-(4-pyridyl)-1,2,4-triazole, FYX-051 - a xanthine oxidoreductase inhibitor for the treatment of hyperuricemia [... Bioorg Med Chem Lett. 2009 Nov 1;19(21):6225-9. | |||||
| REF 33 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2646). | |||||
| REF 34 | DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011 Jan;39(Database issue):D1035-41. | |||||
| REF 35 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 36 | Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate. J Biochem. 2007 Apr;141(4):513-24. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

